| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,499,486 – 9,499,557 |
| Length | 71 |
| Max. P | 0.950854 |

| Location | 9,499,486 – 9,499,557 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Shannon entropy | 0.37177 |
| G+C content | 0.45188 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -16.09 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
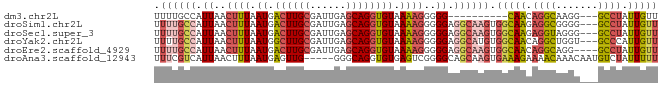
>dm3.chr2L 9499486 71 + 23011544 UUUUGCCAUUAACUUUAAUGACUUGCGAUUGAGCAGGUGUAAAAGGGGG----------CAACAGGCAAGG---GCCUAUUGUU ..(((((.....((((.((.((((((......)))))))).))))..))----------))).((((....---))))...... ( -21.50, z-score = -1.89, R) >droSim1.chr2L 9268785 81 + 22036055 UUUUGCCAUUAACUUUAAUGACUUGCGAUUGAGCAGGUGUAAAAGGGGGAGGCAAGUGGCAAGAGGCGGGG---GCCUAUUGUU .((((((.((..((((.((.((((((......)))))))).))))..)).)))))).(((((.((((....---)))).))))) ( -27.70, z-score = -3.12, R) >droSec1.super_3 4946231 81 + 7220098 UUUUGCCAUUAACUUUAAUGACUUGCGAUUGAGCAGGUGUAAAAGGGGGAGGCAAGUGGCAAGAGGUAGGG---GCCUAUUGUU .((((((.((..((((.((.((((((......)))))))).))))..)).)))))).(((((.((((....---)))).))))) ( -25.80, z-score = -2.75, R) >droYak2.chr2L 12167399 81 + 22324452 UUUUGCCAUUAACUUUAAUGGCUUGCGAUUGAGCAGGUGUAAAAGGGGGAGGCAUGUGGCAACAGGCUGGU---GCCCAUUGUU (((((((((((....)))))((((((......))))))))))))(((.(.(((...((....)).)))..)---.)))...... ( -24.70, z-score = -0.47, R) >droEre2.scaffold_4929 10102441 80 + 26641161 UUUUGCCAUUAACUUUAAUGACUUGCGAUUGAGCAGGUGUAAAAGGGGGAGGCAAGUGGCAACAGGCAGG----GCCUAUUGUU .((((((.((..((((.((.((((((......)))))))).))))..)).)))))).(((((.((((...----)))).))))) ( -26.00, z-score = -2.40, R) >droAna3.scaffold_12943 868695 79 + 5039921 UUUCGUCAUUAACUUUAAUGAGUUG-----GGGCAGGUGUGAGUCGGGGCAGCAAGUGAAAGAAAACAAACAAUGUCUAUUUUU ....(((.(((((((....))))))-----))))............(((((.....((........)).....)))))...... ( -12.20, z-score = 0.70, R) >consensus UUUUGCCAUUAACUUUAAUGACUUGCGAUUGAGCAGGUGUAAAAGGGGGAGGCAAGUGGCAACAGGCAGGG___GCCUAUUGUU .((((((.((..((((.((.((((((......)))))))).))))..)).)))))).(((((.((((.......)))).))))) (-16.09 = -17.60 + 1.51)

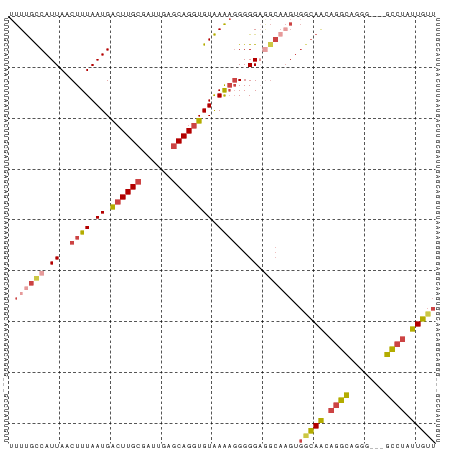
| Location | 9,499,486 – 9,499,557 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Shannon entropy | 0.37177 |
| G+C content | 0.45188 |
| Mean single sequence MFE | -16.36 |
| Consensus MFE | -10.79 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
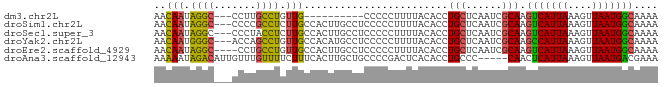
>dm3.chr2L 9499486 71 - 23011544 AACAAUAGGC---CCUUGCCUGUUG----------CCCCCUUUUACACCUGCUCAAUCGCAAGUCAUUAAAGUUAAUGGCAAAA ..((((((((---....))))))))----------..............(((......))).(((((((....))))))).... ( -18.20, z-score = -2.82, R) >droSim1.chr2L 9268785 81 - 22036055 AACAAUAGGC---CCCCGCCUCUUGCCACUUGCCUCCCCCUUUUACACCUGCUCAAUCGCAAGUCAUUAAAGUUAAUGGCAAAA ..(((.((((---....)))).)))....(((((.....((((...((.(((......))).))....)))).....))))).. ( -17.70, z-score = -3.04, R) >droSec1.super_3 4946231 81 - 7220098 AACAAUAGGC---CCCUACCUCUUGCCACUUGCCUCCCCCUUUUACACCUGCUCAAUCGCAAGUCAUUAAAGUUAAUGGCAAAA ..(((.(((.---.....))).)))....(((((.....((((...((.(((......))).))....)))).....))))).. ( -14.70, z-score = -2.13, R) >droYak2.chr2L 12167399 81 - 22324452 AACAAUGGGC---ACCAGCCUGUUGCCACAUGCCUCCCCCUUUUACACCUGCUCAAUCGCAAGCCAUUAAAGUUAAUGGCAAAA ..((((((((---....))))))))........................(((......))).(((((((....))))))).... ( -20.30, z-score = -1.66, R) >droEre2.scaffold_4929 10102441 80 - 26641161 AACAAUAGGC----CCUGCCUGUUGCCACUUGCCUCCCCCUUUUACACCUGCUCAAUCGCAAGUCAUUAAAGUUAAUGGCAAAA ..((((((((----...))))))))....(((((.....((((...((.(((......))).))....)))).....))))).. ( -20.80, z-score = -3.42, R) >droAna3.scaffold_12943 868695 79 - 5039921 AAAAAUAGACAUUGUUUGUUUUCUUUCACUUGCUGCCCCGACUCACACCUGCCC-----CAACUCAUUAAAGUUAAUGACGAAA .(((((((((...)))))))))..(((..(((..((..............))..-----))).((((((....)))))).))). ( -6.44, z-score = 0.34, R) >consensus AACAAUAGGC___CCCUGCCUGUUGCCACUUGCCUCCCCCUUUUACACCUGCUCAAUCGCAAGUCAUUAAAGUUAAUGGCAAAA ..(((.((((.......)))).)))........................(((......))).(((((((....))))))).... (-10.79 = -10.35 + -0.44)
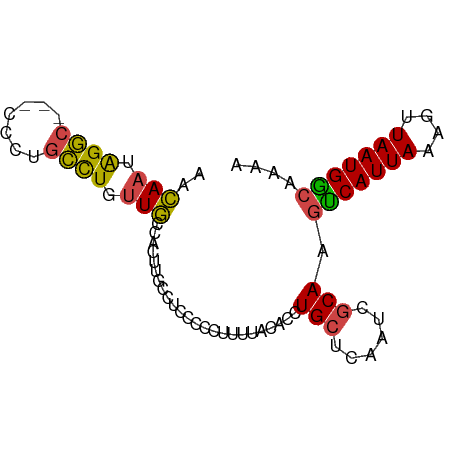
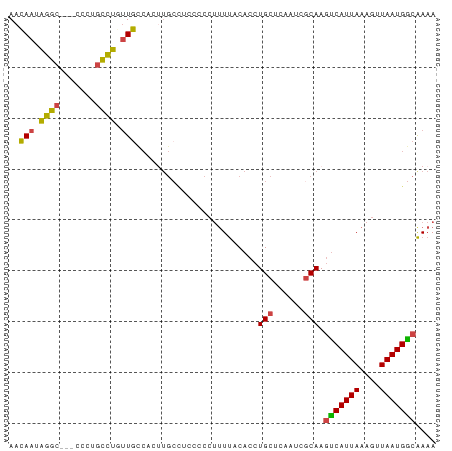
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:26 2011