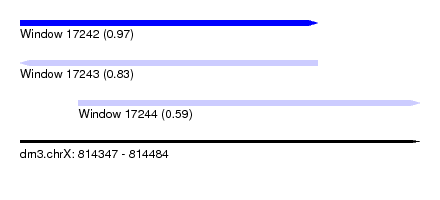
| Sequence ID | dm3.chrX |
|---|---|
| Location | 814,347 – 814,484 |
| Length | 137 |
| Max. P | 0.972833 |

| Location | 814,347 – 814,449 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 90.74 |
| Shannon entropy | 0.18070 |
| G+C content | 0.38164 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 814347 102 + 22422827 -------------CACAUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------............(-----((((((((.....)))))))))((..((((........)))).........(((((((((.....-))))))))))).(((....))).. ( -25.90, z-score = -2.49, R) >droSim1.chrX 651155 102 + 17042790 -------------CACAUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------............(-----((((((((.....)))))))))((..((((........)))).........(((((((((.....-))))))))))).(((....))).. ( -25.90, z-score = -2.49, R) >droSec1.super_10 649332 102 + 3036183 -------------CACAUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUCGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------...........((-----(.((((((.....)))))))))((..((((........)))).........(((((((((.....-))))))))))).(((....))).. ( -25.20, z-score = -2.35, R) >droYak2.chrX 730382 102 + 21770863 -------------CACAUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------............(-----((((((((.....)))))))))((..((((........)))).........(((((((((.....-))))))))))).(((....))).. ( -25.90, z-score = -2.49, R) >droEre2.scaffold_4644 783333 102 + 2521924 -------------CUCGUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGGCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------((((((.......-----((((((((.....))))))))))))))........................(((((((((.....-)))))))))...(((....))).. ( -26.01, z-score = -1.58, R) >droAna3.scaffold_12929 2710726 115 - 3277472 GGAUAGGUAUGUGCACAUAUAUCAGA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGACCGUG-UCGUAAAUUGCAGCCGCCGGGCAG .....(((((((....)))))))..(-----((((((((.....)))))))))((..((((........)))).........((((((((((...)-))))))))))).(((....))).. ( -30.10, z-score = -2.24, R) >droWil1.scaffold_181096 1615582 112 - 12416693 ---------CGUAUUCGUAUAUAGAAGAGAAAAUAUGCUAAAUCAGCAUAUUUGUGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGACCCAGGCCGUAAAUUGCAGCCGCCGGGCAG ---------.((....(((.(((((.....(((((((((.....))))))))).....))))).)))....)).........((((((((.(....).))))))))...(((....))).. ( -26.20, z-score = -2.18, R) >droMoj3.scaffold_6473 6635806 102 + 16943266 -------------UUCAUAUAUCACA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------........((.((-----((((((((.....)))))))))).))((((........)))).........((((((((((...)-)))))))))...(((....))).. ( -29.70, z-score = -3.65, R) >droVir3.scaffold_12726 978394 102 - 2840439 -------------UUCAUAUAUCACA-----AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCCGUG-CCGUAAAUUGCAGCCGCCGGGCAG -------------........((.((-----((((((((.....)))))))))).))((((........)))).........((((((((((...)-)))))))))...(((....))).. ( -29.70, z-score = -3.65, R) >droGri2.scaffold_15203 11202300 102 - 11997470 -------------UUCAUAUAUCAUA-----AAUAUGCUAAAUCAGUAUAUUUGUGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCCCUG-CCGUAAAUUUCCGCCGCCGGGCAG -------------........(((((-----((((((((.....)))))))))))))............(((..........((((((((((...)-))))))))))))(((....))).. ( -30.20, z-score = -4.47, R) >consensus _____________CACAUAUAUCAGA_____AAUAUGCUAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCCGUG_CCGUAAAUUGCAGCCGCCGGGCAG ...............................((((((((.....))))))))(((..((((........)))).........(((((((((......))))))))))))(((....))).. (-21.35 = -21.42 + 0.07)
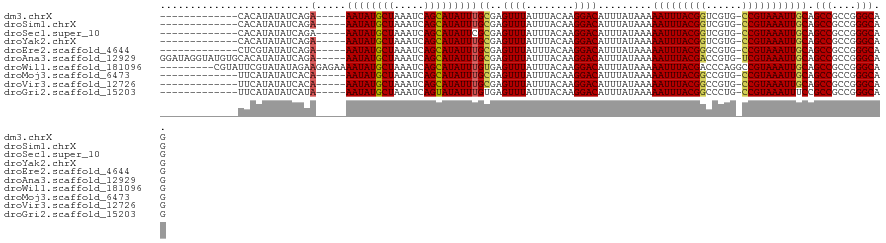
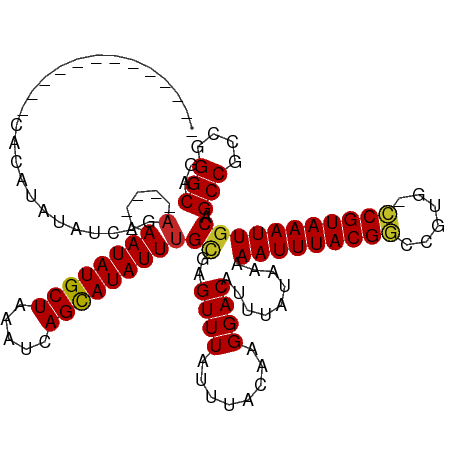
| Location | 814,347 – 814,449 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 90.74 |
| Shannon entropy | 0.18070 |
| G+C content | 0.38164 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 814347 102 - 22422827 CUGCCCGGCGGCUGCAAUUUACGG-CACGACCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUAUGUG------------- ..(((....))).(((((((((((-.....)))))))))(((((((......))))))).......))((((((((((...)))))))))-----)............------------- ( -23.50, z-score = -1.59, R) >droSim1.chrX 651155 102 - 17042790 CUGCCCGGCGGCUGCAAUUUACGG-CACGACCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUAUGUG------------- ..(((....))).(((((((((((-.....)))))))))(((((((......))))))).......))((((((((((...)))))))))-----)............------------- ( -23.50, z-score = -1.59, R) >droSec1.super_10 649332 102 - 3036183 CUGCCCGGCGGCUGCAAUUUACGG-CACGACCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCGAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUAUGUG------------- ..(((....))).(((((((((((-.....)))))))))(((((((......))))))).......))((((((((((...)))))))))-----)............------------- ( -23.90, z-score = -1.51, R) >droYak2.chrX 730382 102 - 21770863 CUGCCCGGCGGCUGCAAUUUACGG-CACGACCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUAUGUG------------- ..(((....))).(((((((((((-.....)))))))))(((((((......))))))).......))((((((((((...)))))))))-----)............------------- ( -23.50, z-score = -1.59, R) >droEre2.scaffold_4644 783333 102 - 2521924 CUGCCCGGCGGCUGCAAUUUACGG-CACGCCCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUACGAG------------- ......((((((((.......)))-).))))((((....(((((((......))))))).....(((.((((((((((...)))))))))-----).)))..))))..------------- ( -26.90, z-score = -2.49, R) >droAna3.scaffold_12929 2710726 115 + 3277472 CUGCCCGGCGGCUGCAAUUUACGA-CACGGUCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UCUGAUAUAUGUGCACAUACCUAUCC ..(((....))).(((((((((((-(...))))))))))(((((((......))))))).......))((((((((((...)))))))))-----)..((((((((....))))..)))). ( -26.20, z-score = -1.87, R) >droWil1.scaffold_181096 1615582 112 + 12416693 CUGCCCGGCGGCUGCAAUUUACGGCCUGGGUCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCACAAAUAUGCUGAUUUAGCAUAUUUUCUCUUCUAUAUACGAAUACG--------- ..(((....)))...((((((((((....)))))))))).........((..(((((.(((.......((((((((((...)))))))))).......))).)))))..)).--------- ( -26.04, z-score = -2.56, R) >droMoj3.scaffold_6473 6635806 102 - 16943266 CUGCCCGGCGGCUGCAAUUUACGG-CACGGCCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UGUGAUAUAUGAA------------- ..(((....)))...(((((((((-(...))))))))))(((((((......))))))).....((((((((((((((...)))))))))-----)))))........------------- ( -31.90, z-score = -3.43, R) >droVir3.scaffold_12726 978394 102 + 2840439 CUGCCCGGCGGCUGCAAUUUACGG-CACGGCCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU-----UGUGAUAUAUGAA------------- ..(((....)))...(((((((((-(...))))))))))(((((((......))))))).....((((((((((((((...)))))))))-----)))))........------------- ( -31.90, z-score = -3.43, R) >droGri2.scaffold_15203 11202300 102 + 11997470 CUGCCCGGCGGCGGAAAUUUACGG-CAGGGCCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCACAAAUAUACUGAUUUAGCAUAUU-----UAUGAUAUAUGAA------------- (((((....)))))((((((((((-(...)))))))))))((((((......))))))......(((.((((((.(((...))).)))))-----).)))........------------- ( -23.40, z-score = -1.41, R) >consensus CUGCCCGGCGGCUGCAAUUUACGG_CACGGCCGUAAAUUUUUAUAAAUGUCCUUGUAAAUAAACUCGCAAAUAUGCUGAUUUAGCAUAUU_____UCUGAUAUAUGAG_____________ ..(((....)))((((((((((((......)))))))))(((((((......))))))).......)))(((((((((...)))))))))............................... (-20.18 = -20.25 + 0.07)

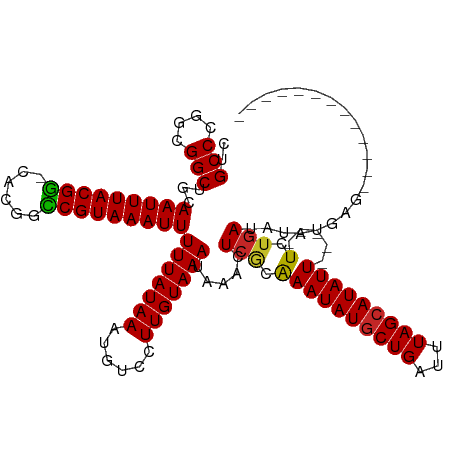
| Location | 814,367 – 814,484 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Shannon entropy | 0.11453 |
| G+C content | 0.39022 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.21 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 814367 117 + 22422827 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAGAA ..((((.(((....)))...............(((.........(((((((((..-...)))))))))((((((....)))(((((....))))).))))--))...))))......... ( -28.30, z-score = -0.93, R) >droSim1.chrX 651175 117 + 17042790 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAGAA ..((((.(((....)))...............(((.........(((((((((..-...)))))))))((((((....)))(((((....))))).))))--))...))))......... ( -28.30, z-score = -0.93, R) >droSec1.super_10 649352 117 + 3036183 UAAAUCAGCAUAUUCGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAGAA .........(((.((((((((..((((.((((..(.........(((((((((..-...)))))))))...(((....))).)..))))))))....)).--))).))).)))....... ( -28.90, z-score = -1.07, R) >droYak2.chrX 730402 117 + 21770863 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGUC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGUG--UCGACGAUUAUAUCGGAA ((((((.(((....)))...........((((..(.........(((((((((..-...)))))))))...(((....))).)..)))).))))))....--....((((...))))... ( -28.60, z-score = -0.81, R) >droEre2.scaffold_4644 783353 117 + 2521924 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGGC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAGAA ..((((.(((....)))...............(((.........(((((((((..-...)))))))))((((((....)))(((((....))))).))))--))...))))......... ( -27.50, z-score = -0.44, R) >droAna3.scaffold_12929 2710759 117 - 3277472 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGACC-GUGUCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUACCAAAA ..((((.(((....)))...............(((.........((((((((((.-..))))))))))((((((....)))(((((....))))).))))--))...))))......... ( -27.30, z-score = -1.16, R) >dp4.chrXL_group3a 2656791 117 + 2690836 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC-GUGCCGUAAAUUGCAGCCUCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAAAA ..((((.(((....)))...............(((.........((((((((((.-..))))))))))((((((....)))(((((....))))).))))--))...))))......... ( -29.60, z-score = -1.65, R) >droPer1.super_11 825135 117 - 2846995 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC-GUGCCGUAAAUUGCAGCCUCCGGGCAGGUCUUGAGAUUUAUGCG--UCGACGAUUAUAUCAAAA ..((((.(((....)))...............(((.........((((((((((.-..))))))))))((((((....)))(((((....))))).))))--))...))))......... ( -29.60, z-score = -1.65, R) >droWil1.scaffold_181096 1615611 120 - 12416693 UAAAUCAGCAUAUUUGUGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGACCCAGGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUAUGCCUCGACGAUUAUAUCAAAA .......((((((.((((((....))))))(((((.........((((((((.(....).))))))))...(((....)))..))))).......))))))................... ( -24.20, z-score = -0.47, R) >droMoj3.scaffold_6473 6635826 117 + 16943266 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCC--UCGCCGAUUAUAUCAAAA ..((((.(((....)))(((..((..(...(((((.........((((((((((.-..))))))))))...(((....)))..)))))...)..))...)--))...))))......... ( -29.10, z-score = -1.01, R) >droVir3.scaffold_12726 978414 117 - 2840439 UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC-GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCC--UCGCCGAUUAUAUCAAAA ..((((.(((....)))(((..((..(...(((((.........((((((((((.-..))))))))))...(((....)))..)))))...)..))...)--))...))))......... ( -29.10, z-score = -1.01, R) >droGri2.scaffold_15203 11202320 117 - 11997470 UAAAUCAGUAUAUUUGUGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC-CUGCCGUAAAUUUCCGCCGCCGGGCAGAUCUAGAGAUUUAUACC--UCGCCGAUUAUAUGAAAA .......((...((((((((....)))))))).)).((((((.(((((((((((.-..)))))))))))..(((....))).((((..(((........)--))...)))).)))))).. ( -28.10, z-score = -2.18, R) >consensus UAAAUCAGCAUAUUUGCGAGUUUAUUUACAAGGACAUUUAUAAAAAUUUACGGCC_GUGCCGUAAAUUGCAGCCGCCGGGCAGGUCUUGAGAUUUAUGCG__UCGACGAUUAUAUCAAAA ((((((.(((....)))...........((((..(.........((((((((((....))))))))))...(((....))).)..)))).))))))........................ (-26.42 = -26.21 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:13 2011