| Sequence ID | dm3.chrX |
|---|---|
| Location | 761,443 – 761,508 |
| Length | 65 |
| Max. P | 0.973562 |

| Location | 761,443 – 761,508 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Shannon entropy | 0.10440 |
| G+C content | 0.43506 |
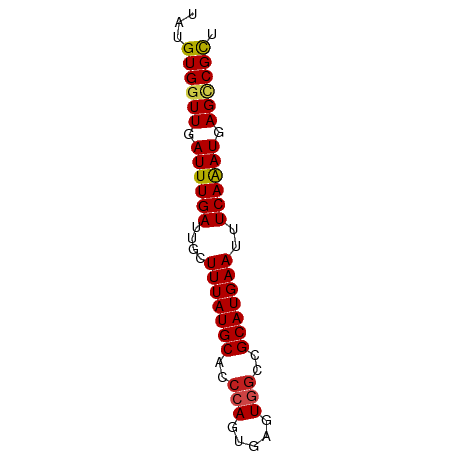
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
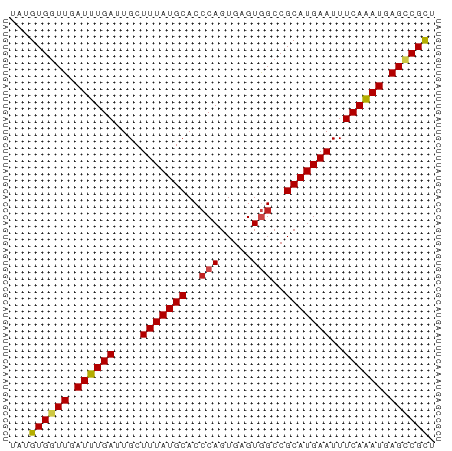
>dm3.chrX 761443 65 + 22422827 UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGCCGUU .....((((.((((((....(((((((..(((.....)))..)))))))..)))))).))))... ( -20.50, z-score = -2.78, R) >droSim1.chrX_random 822632 65 + 5698898 UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGUCGUU .....(..(.((((((....(((((((..(((.....)))..)))))))..)))))).)..)... ( -16.20, z-score = -1.19, R) >droSec1.super_10 598291 65 + 3036183 UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGUCGUU .....(..(.((((((....(((((((..(((.....)))..)))))))..)))))).)..)... ( -16.20, z-score = -1.19, R) >droYak2.chrX 678559 65 + 21770863 UAUGUGGUUGAUUUGAUUGCUUUAUGCACACAGUGAGUAGCCGCAUGAAUUUCAAAUGAGUCGUU ((((((((((..(((..(((.....)))..)))....)))))))))).................. ( -15.40, z-score = -0.90, R) >droEre2.scaffold_4644 722925 65 + 2521924 UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUAGCCGCAUGAAUUUCAAAUGAGGCGUU ((((((((((..(((..(((.....)))..)))....)))))))))).................. ( -16.30, z-score = -1.24, R) >droAna3.scaffold_12929 510864 64 + 3277472 UUUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGUCGC- ...(((..(.((((((....(((((((..(((.....)))..)))))))..)))))).)..)))- ( -18.40, z-score = -1.95, R) >dp4.chrXL_group3a 2557290 65 + 2690836 UUUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAGAUGAGCCGCU ...((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). ( -24.00, z-score = -3.77, R) >droPer1.super_11 719296 65 - 2846995 UUUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAGAUGAGCCGCU ...((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). ( -24.00, z-score = -3.77, R) >droWil1.scaffold_181096 1537102 64 - 12416693 -UUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGACGCC -..(((.((.((((((....(((((((..(((.....)))..)))))))..)))))).)).))). ( -16.60, z-score = -1.45, R) >droVir3.scaffold_12726 902995 64 - 2840439 -UUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGCCGCU -..((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). ( -23.90, z-score = -3.83, R) >droMoj3.scaffold_6473 6547611 65 + 16943266 UUUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGCCGCU ...((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). ( -23.90, z-score = -3.81, R) >droGri2.scaffold_15203 11108288 65 - 11997470 UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGCCGCU ...((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). ( -23.90, z-score = -3.80, R) >consensus UAUGUGGUUGAUUUGAUUGCUUUAUGCACCCAGUGAGUGGCCGCAUGAAUUUCAAAUGAGCCGCU ...((((((.((((((....(((((((..(((.....)))..)))))))..)))))).)))))). (-17.86 = -17.73 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:02 2011