
| Sequence ID | dm3.chrX |
|---|---|
| Location | 760,025 – 760,126 |
| Length | 101 |
| Max. P | 0.791603 |

| Location | 760,025 – 760,119 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 63.05 |
| Shannon entropy | 0.62598 |
| G+C content | 0.43092 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -8.08 |
| Energy contribution | -11.80 |
| Covariance contribution | 3.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
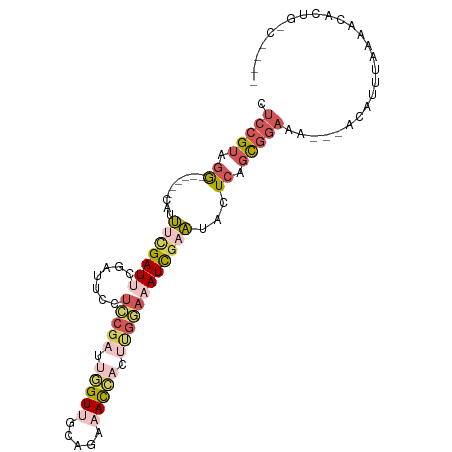
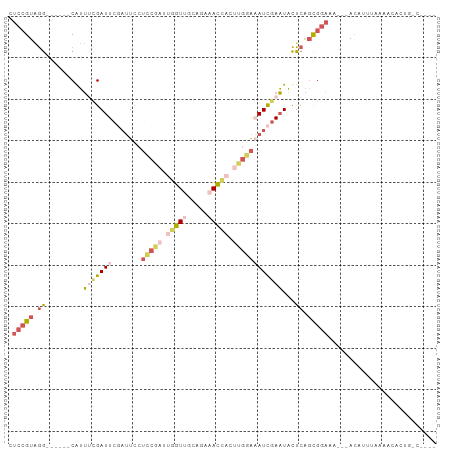
>dm3.chrX 760025 94 - 22422827 CUCCGUAGG------CAUUACGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUUAAAACACUGUCUCCU .(((((.(.------.....).((((((((..(((((.(((((.....))))).))))))))))))).....)))))..---..................... ( -24.80, z-score = -2.71, R) >droWil1.scaffold_181096 10880852 84 - 12416693 UACCGUUAA------CAAGCAGAUUCUCCUCCAGCUCUCAGU-ACACUUAUUUUUUAUGCAUU-AUUUUUCGUUCUCCGUUGAUGAUUGGCA----------- ...((((((------(.(((.((......))..)))....((-(.............)))...-..............))))))).......----------- ( -6.32, z-score = 1.54, R) >droSim1.chrX_random 820790 90 - 5698898 CUCCGUAGG------CGUUUCGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUAAAAACACUGGC---- .(((((.((------.((....((((((((..(((((.(((((.....))))).))))))))))))))))).)))))..---.................---- ( -25.70, z-score = -2.39, R) >droSec1.super_10 596900 90 - 3036183 CUCCGUAGG------CGUUUCGAUUCUAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUAAAAACACUGGC---- .(((((.((------.(((((((((.......(((((.(((((.....))))).)))))))))))).)))).)))))..---.................---- ( -24.51, z-score = -2.44, R) >droYak2.chrX 677172 96 - 21770863 CUCCGUGGGGCUUCACAUUUUGAUUCGAUUGCUACGAUUGGUGGCAGAAACACCCCGAAUAUCGAAUACUCAGCGGAAAAUUACGUUUAAAACACA------- .(((((((((..(((.....))).....(((((((.....))))))).....)))).......((....)).)))))...................------- ( -21.50, z-score = -0.72, R) >consensus CUCCGUAGG______CAUUUCGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA___ACAUUUAAAACACUG_C____ .(((((.((.............((((((((..(((((.(((((.....))))).)))))))))))))..)).))))).......................... ( -8.08 = -11.80 + 3.72)

| Location | 760,032 – 760,126 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.55706 |
| G+C content | 0.43504 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -11.48 |
| Energy contribution | -13.32 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
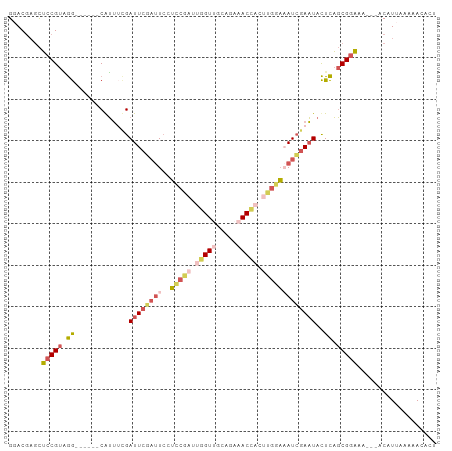
>dm3.chrX 760032 94 - 22422827 GGACGAGCUCCGUAGG------CAUUACGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUUAAAACACU ........(((((.(.------.....).((((((((..(((((.(((((.....))))).))))))))))))).....)))))..---.............. ( -24.80, z-score = -2.28, R) >droWil1.scaffold_181096 10880856 87 - 12416693 GUACCUUUACCGUUAA------CAAGCAGAUUCUCCUCCAGCUCUCAGU-----ACACUUAUUUUUUAUGCAUUAUUUUUCGUUCU---CCGUUGAUGAUU-- ..........((((((------(.(((.((......))..)))....((-----(.............)))...............---..)))))))...-- ( -6.32, z-score = 0.58, R) >droSim1.chrX_random 820793 94 - 5698898 GGACGAGCUCCGUAGG------CGUUUCGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUAAAAACACU ........(((((.((------.((....((((((((..(((((.(((((.....))))).))))))))))))))))).)))))..---.............. ( -25.70, z-score = -1.80, R) >droSec1.super_10 596903 94 - 3036183 GGACGAGCUCCGUAGG------CGUUUCGAUUCUAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA---ACAUUAAAAACACU ........(((((.((------.(((((((((.......(((((.(((((.....))))).)))))))))))).)))).)))))..---.............. ( -24.51, z-score = -1.82, R) >droYak2.chrX 677172 103 - 21770863 GCACGAGCUCCGUGGGGCUUCACAUUUUGAUUCGAUUGCUACGAUUGGUGGCAGAAACACCCCGAAUAUCGAAUACUCAGCGGAAAAUUACGUUUAAAACACA ((....))(((((((((..(((.....))).....(((((((.....))))))).....)))).......((....)).)))))................... ( -22.00, z-score = -0.05, R) >consensus GGACGAGCUCCGUAGG______CAUUUCGAUUCGAUUCCUCCGAUUGGUUGCAGAAACCACUUGGAAAUCGAAUACUCAGCGGAAA___ACAUUAAAAACACU ........(((((.((.............((((((((..(((((.(((((.....))))).)))))))))))))..)).)))))................... (-11.48 = -13.32 + 1.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:04:01 2011