
| Sequence ID | dm3.chrX |
|---|---|
| Location | 732,788 – 732,872 |
| Length | 84 |
| Max. P | 0.582763 |

| Location | 732,788 – 732,872 |
|---|---|
| Length | 84 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 63.27 |
| Shannon entropy | 0.67270 |
| G+C content | 0.43383 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 732788 84 - 22422827 ----UCUAUCCCGA----ACACAACGGUCUUUUCGUGUAGGGUAUUUGCGGUGCCGUUCUCUCUUCUUCAGUCUUUUUGAGAGGGCACUUUG------- ----.....(((..----((((............)))).))).......((((((...(((((...............)))))))))))...------- ( -20.66, z-score = -0.93, R) >droSim1.chrX 581761 84 - 17042790 ----UCUAUCCCCA----ACAUAAAGAUCUUUUCGUGUAGGGUAUUUGCGGUGCCGUUCUCUCUUCUUCAGUCUUUUCGAGAGGGCACUUUG------- ----......(((.----(((..(((....)))..))).))).......((((((...(((((...............)))))))))))...------- ( -20.26, z-score = -1.49, R) >droSec1.super_10 570033 84 - 3036183 ----UCUAUCCCCA----ACAUAACGAUCUUUUCGUGUAGGGUAUUUGCGGUGCCGUUCUCUCUUCUUCAGUUUUUUCGAGAGGGCACUUUG------- ----......(((.----((...((((.....)))))).))).......((((((...(((((...............)))))))))))...------- ( -20.66, z-score = -1.77, R) >droYak2.chrX 648371 84 - 21770863 ----UCUAUCCGCA----ACACAAAGAUCUUUUCGUGUAGGGUAUUUGCAGUGCCGUUCCCUCUUCCCCUGACUUUUUGAGAGGGCUCUUUG------- ----..........----...((((((.............(((((.....)))))...(((((((.............))))))).))))))------- ( -17.62, z-score = -0.07, R) >droEre2.scaffold_4644 692401 88 - 2521924 UCUACAUAUCUCCA----ACAUAACGAUCUUCUCGUGUAGGGUAUUUGCAGUGCCGUUCUCUCUUCUUCAGUCUUUUUGAGAGGGCUCUUUG------- .((((.(((.....----..)))((((.....))))))))(((((.....)))))((((((((...............))))))))......------- ( -18.46, z-score = -1.16, R) >droAna3.scaffold_12929 471524 82 - 3277472 ----UCUACCCUAAAACUAUAUAAAGGCACCUUCUUCUAGAGUAUUUUCUUAGUCGCUACCCUAUUAUUUCCCUU------AGUGCAUUUCG------- ----......................((((........((((....))))(((........)))...........------.))))......------- ( -5.30, z-score = 0.89, R) >droGri2.scaffold_15203 11056319 89 + 11997470 ----UCUAUGCG------UCGUAUUGGACCAAUCGAGUAAACCAUUUUCCGUUUUUCCGUUAUAGCCCGUUUUUCACAAUGAUUGUGCAUAAAACUUCA ----..((((((------(..((.((((..(((.(((.........))).)))..)))).))..)).((((......)))).....)))))........ ( -12.00, z-score = 0.06, R) >consensus ____UCUAUCCCCA____ACAUAAAGAUCUUUUCGUGUAGGGUAUUUGCGGUGCCGUUCUCUCUUCUUCAGUCUUUUUGAGAGGGCACUUUG_______ .......................((((.....))))....(((((.....)))))((((((((...............))))))))............. ( -7.85 = -8.20 + 0.36)

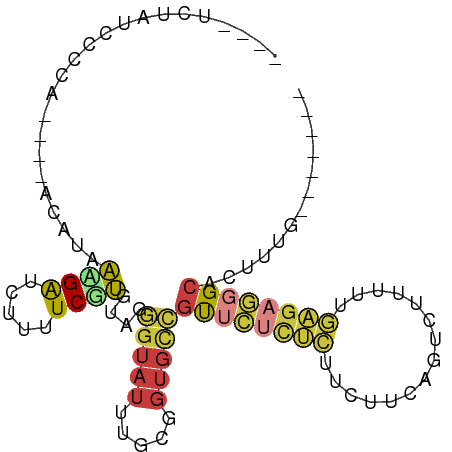

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:57 2011