| Sequence ID | dm3.chrX |
|---|---|
| Location | 697,025 – 697,140 |
| Length | 115 |
| Max. P | 0.844496 |

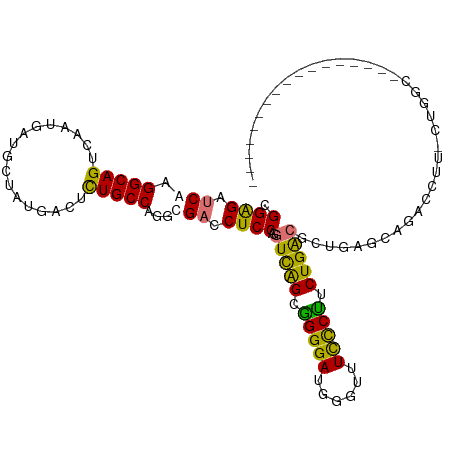
| Location | 697,025 – 697,140 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.46652 |
| G+C content | 0.58907 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -26.83 |
| Energy contribution | -25.96 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
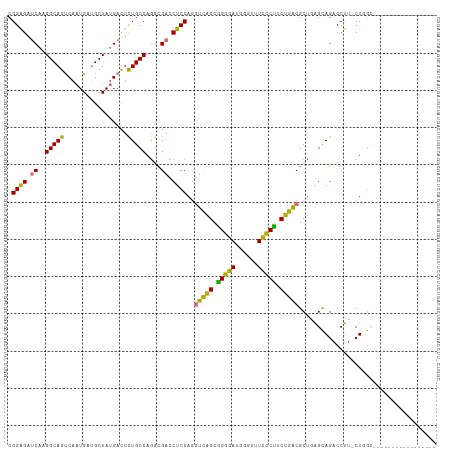
>dm3.chrX 697025 115 + 22422827 CGGAGAGCAAGGCAGUCAAUGAUGCUAUGACUCUGCCAAGUGACCUCCAGGUCGGCGGGGAUUGGUGUUCCCUCUGACGCCAAGCAGCCCUGACUCGUCAUUACUUGUUUCUGGC ((((((.(((((((((((....((((....(((((((....((((....)))))))))))..(((((((......)))))))))))....))))).)).....)))))))))).. ( -42.30, z-score = -1.10, R) >droSec1.super_10 547595 97 + 3036183 CGGAGAUCAAGGCAGUCAAUGCUGCUAUGAUUCUGCCAGGCGACCUCCAUGUCGGCGGGGAUGGGUUUCCCUUCUGACGCUGAGCAGACCUU-CUGGC----------------- (((((((((.((((((....)))))).))))(((((..((((.....)..(((((.(((((......))))).))))))))..)))))..))-)))..----------------- ( -39.70, z-score = -1.62, R) >droSim1.chrX 570236 97 + 17042790 CGGAGAUCAAGGCAGUCAAUGCUGCUAUGACUCUGCCAGGCGACCUCCAGGUCAGCGGGGAUGGGUUUCCCUUCUGACGCUGAGCAGACCUU-CUGGC----------------- .((((.(((.((((((....)))))).)))))))((((((.(..(((((((((((.(((((......))))).))))).))).).))..).)-)))))----------------- ( -43.70, z-score = -2.42, R) >droEre2.scaffold_4644 666830 109 + 2521924 CGGGGAUCACGGCAGCCGGUGAGGCUAGGACUUUGCCAGAUGACCUCCAGCUUAGCCGGGAUUGGUUUCUCGUCUGGCGCUUGGUAGUCCCAACUUAGGCGUUCCUGGG------ ..(((((.(((((((((.....))))........(((((((((...((((((......)).))))....))))))))))))..)).)))))..((((((....))))))------ ( -42.00, z-score = -0.23, R) >consensus CGGAGAUCAAGGCAGUCAAUGAUGCUAUGACUCUGCCAGGCGACCUCCAGGUCAGCGGGGAUGGGUUUCCCUUCUGACGCUGAGCAGACCUU_CUGGC_________________ .((((.(((.(((((.................)))))...))).))))..(((((.(((((......))))).)))))..................................... (-26.83 = -25.96 + -0.87)

| Location | 697,025 – 697,140 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.46652 |
| G+C content | 0.58907 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
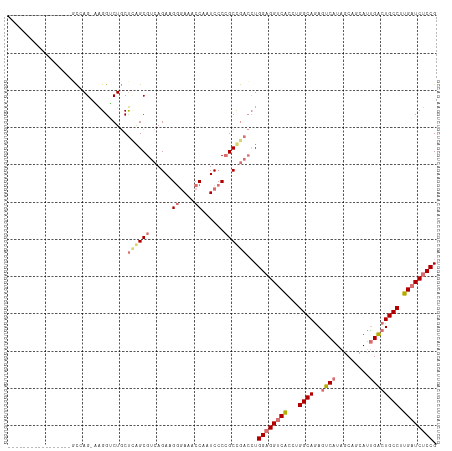
>dm3.chrX 697025 115 - 22422827 GCCAGAAACAAGUAAUGACGAGUCAGGGCUGCUUGGCGUCAGAGGGAACACCAAUCCCCGCCGACCUGGAGGUCACUUGGCAGAGUCAUAGCAUCAUUGACUGCCUUGCUCUCCG ...................(((.((((((((((.((((.....((((.......))))))))((((....))))....)))).(((((.........)))))))))))))).... ( -37.20, z-score = -0.43, R) >droSec1.super_10 547595 97 - 3036183 -----------------GCCAG-AAGGUCUGCUCAGCGUCAGAAGGGAAACCCAUCCCCGCCGACAUGGAGGUCGCCUGGCAGAAUCAUAGCAGCAUUGACUGCCUUGAUCUCCG -----------------(((((-..(..((.(.((..(((.(..((((......))))..).))).)))))..)..)))))(((.(((..((((......))))..))))))... ( -33.00, z-score = -1.04, R) >droSim1.chrX 570236 97 - 17042790 -----------------GCCAG-AAGGUCUGCUCAGCGUCAGAAGGGAAACCCAUCCCCGCUGACCUGGAGGUCGCCUGGCAGAGUCAUAGCAGCAUUGACUGCCUUGAUCUCCG -----------------(((((-..(..((.(.(((.(((((..((((......))))..)))))))))))..)..))))).((((((..((((......))))..))).))).. ( -42.80, z-score = -3.27, R) >droEre2.scaffold_4644 666830 109 - 2521924 ------CCCAGGAACGCCUAAGUUGGGACUACCAAGCGCCAGACGAGAAACCAAUCCCGGCUAAGCUGGAGGUCAUCUGGCAAAGUCCUAGCCUCACCGGCUGCCGUGAUCCCCG ------....(((.(((.....((((.....))))((((((((.((....((....(((((...))))).)))).))))))........((((.....)))))).))).)))... ( -33.10, z-score = -0.14, R) >consensus _________________GCCAG_AAGGUCUGCUCAGCGUCAGAAGGGAAACCAAUCCCCGCCGACCUGGAGGUCACCUGGCAGAGUCAUAGCAGCAUUGACUGCCUUGAUCUCCG ................................((((((......((....))......))))))...((((((((...((((..((((.........)))))))).)))))))). (-22.06 = -23.50 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:53 2011