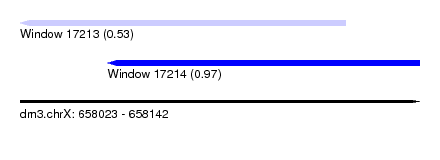
| Sequence ID | dm3.chrX |
|---|---|
| Location | 658,023 – 658,142 |
| Length | 119 |
| Max. P | 0.966487 |

| Location | 658,023 – 658,120 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.46603 |
| G+C content | 0.44293 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
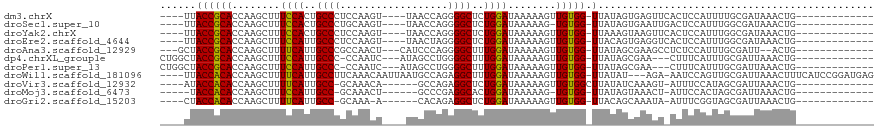
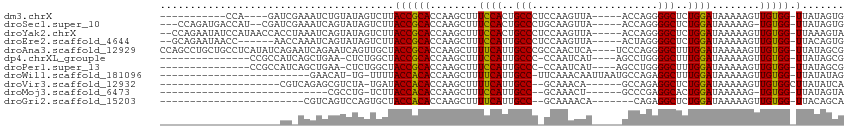
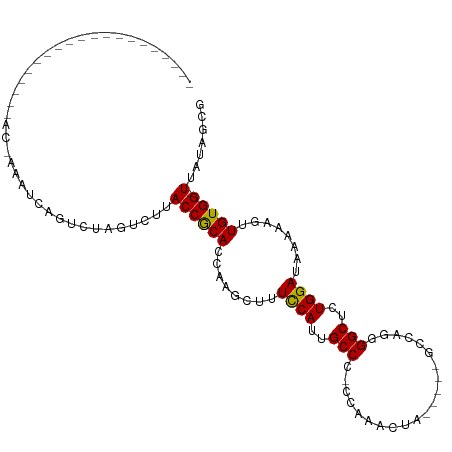
>dm3.chrX 658023 97 - 22422827 ----UUACCGCACCAAGCUUUCCACUGCCCUCCAAGU----UAACCAGGGGCUCUGGAUAAAAAGUUGUGG-UUAUAGUGAGUUCACUCCAUUUUGCGAUAAACUG------------- ----....((((((((((((((((..(((((......----......)))))..))))....))))).)))-)...((((....)))).......)))........------------- ( -25.60, z-score = -1.50, R) >droSec1.super_10 507594 96 - 3036183 ----UUACCGCACCAAGCUUUCCACUGCCCUGCAAGU----UAACCAGGGGCUCUGGAUAAAAAG-UGUGG-UUAUAGUGAAUUGACUCCAUUUGGCGAUAAACUG------------- ----..(((((((.......((((..(((((......----......)))))..))))......)-)))))-)..((((..((((.((......))))))..))))------------- ( -26.82, z-score = -1.35, R) >droYak2.chrX 584422 97 - 21770863 ----UUACCGCACCAAGCUUUCCACUGCCCUCCAAGU----UAACCAGGGGCUCUGGAUAAAAAGUUGUGG-UUAAAGUAAGUUCACUCCAUUUGGCGAUAAACUG------------- ----....((((((((((((((((..(((((......----......)))))..))))....))))).)))-)((((...((....))...)))))))........------------- ( -22.30, z-score = -0.26, R) >droEre2.scaffold_4644 623079 97 - 2521924 ----UUACCGCACCAAGCUUUCCAUUGCCCUCCAAGU----UAACUAGGGGCUCUGGAUAAAAAGUUGUGG-UUACAGUGAGGUCACUCCAUUUGGCGAUAAACUG------------- ----..((((((....((((((((..(((((...((.----...)).)))))..))))....)))))))))-)..((((...(((.((......)).)))..))))------------- ( -27.20, z-score = -1.21, R) >droAna3.scaffold_12929 845442 97 + 3277472 ---GCUACCGCACCAAGCUUUUCAUUGCCCGCCAACU---CAUCCCAGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCGAAGCCUCUCCAUUUGCGAUU--ACUG------------- ---(((..((((((((((((((((..((((.......---........))))..))))....))))).)))-)....))).)))................--....------------- ( -20.66, z-score = 0.59, R) >dp4.chrXL_group1e 7626300 98 - 12523060 CUGGCUACCGCACCAAGCUUUCCAUUGCCC-CCAAUC---AUAGCCUGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCGAA---CUUUCAUUUGCGAUUAAACUG------------- ......((((((....((((((((..((((-(.....---.......)))))..))))....)))))))))-)....(((((---......)))))..........------------- ( -26.30, z-score = -1.11, R) >droPer1.super_13 1344818 98 + 2293547 CUGGCUACCGCACCAAGCUUUCCAUUGCCC-CCAAUC---AUAGCCUGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCGAA---CUUUCAUUUGCGAUUAAACUG------------- ......((((((....((((((((..((((-(.....---.......)))))..))))....)))))))))-)....(((((---......)))))..........------------- ( -26.30, z-score = -1.11, R) >droWil1.scaffold_181096 3326878 110 + 12416693 ----UUACCACACCAAGCUUUUCAUUGCCUUCAAACAAUUAAUGCCAGAGGCUUUGGAUAAAAAGUUGUGG-UUAUAU---AGA-AAUCCAGUUGCGAUUAAACUUUCAUCCGGAUGAG ----..((((((....((((((((..(((((....((.....))...)))))..))))....)))))))))-).....---...-.((((.(.((.((......)).)).).))))... ( -19.40, z-score = 0.06, R) >droVir3.scaffold_12932 1537721 94 - 2102469 ----AUACCACACCAAGCUUUUCAUUGCC-GCAAACA------GCCAGAGGCUCUGGAUAAAAAGUUGUGGCUUAUAUCAAAGU-AUUUCCAUAGCGAUUAAACUG------------- ----........((((((((((..(((..-.....))------)..))))))).))).......(((((((...((((....))-))..)))))))..........------------- ( -18.50, z-score = -0.64, R) >droMoj3.scaffold_6473 13871618 91 + 16943266 -----UACCACACCAAGCUUUCCAUUGCC-GCAAACU------GCCCGAGGCACUGGAUAAAAAG-UGUGG-UUAUAGUAAACU-AUUCCACUAGCGAUUAAACUG------------- -----.(((((((.......((((.((((-((.....------))....)))).))))......)-)))))-).((((....))-))...................------------- ( -22.12, z-score = -2.12, R) >droGri2.scaffold_15203 6328932 92 + 11997470 ----CUACCACACCAAGCUUUUCAUUGCC-GCAAA-A------CACAGAGGCUCUGGAUAAAAAGUUGUGG-UUACAGCAAAUA-AUUUCGGUAGCGAUUAAACUG------------- ----(((((...((((((((((...((..-.....-.------)).))))))).))).......(((((..-..))))).....-.....)))))...........------------- ( -18.30, z-score = -0.47, R) >consensus ____UUACCGCACCAAGCUUUCCAUUGCCCUCCAACU____UAGCCAGGGGCUCUGGAUAAAAAGUUGUGG_UUAUAGCGAAGU_ACUCCAUUUGCGAUUAAACUG_____________ .......(((((........((((..((((..................))))..))))........)))))................................................ (-15.45 = -15.14 + -0.30)
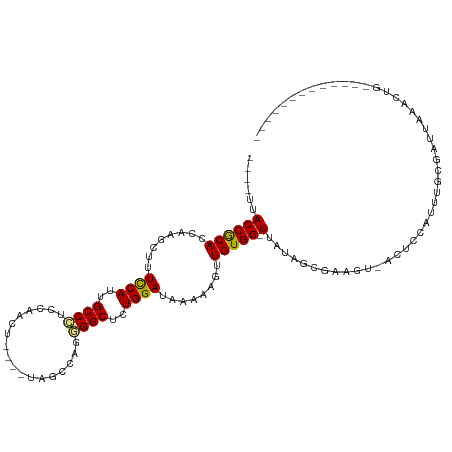
| Location | 658,049 – 658,142 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.68308 |
| G+C content | 0.46599 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 658049 93 - 22422827 -----------CCA----GAUCGAAAUCUGUAUAGUCUUACCGCACCAAGCUUUCCACUGCCCUCCAAGUUA-----ACCAGGGGCUCUGGAUAAAAAGUUGUGG-UUAUAGUG -----------.((----(((....)))))......((.((((((....((((((((..(((((........-----....)))))..))))....)))))))))-)...)).. ( -23.60, z-score = -1.33, R) >droSec1.super_10 507620 102 - 3036183 ---CCAGAUGACCAU--CGAUCGAAAUCAGUAUAGUCUUACCGCACCAAGCUUUCCACUGCCCUGCAAGUUA-----ACCAGGGGCUCUGGAUAAAAAG-UGUGG-UUAUAGUG ---...(((....))--).........((.((((.....(((((((.......((((..(((((........-----....)))))..))))......)-)))))-))))).)) ( -25.82, z-score = -0.97, R) >droYak2.chrX 584448 106 - 21770863 --CCAGAAUAUCCAUAACCACCUAAAUCAGUAUAGUCUUACCGCACCAAGCUUUCCACUGCCCUCCAAGUUA-----ACCAGGGGCUCUGGAUAAAAAGUUGUGG-UUAAAGUA --............(((((((........(((......))).......(((((((((..(((((........-----....)))))..))))....)))))))))-)))..... ( -23.50, z-score = -1.11, R) >droEre2.scaffold_4644 623105 100 - 2521924 --GCAGAAUAACC------AACCAAAUCAGUAUAGUCUUACCGCACCAAGCUUUCCAUUGCCCUCCAAGUUA-----ACUAGGGGCUCUGGAUAAAAAGUUGUGG-UUACAGUG --...........------...............((...((((((....((((((((..(((((...((...-----.)).)))))..))))....)))))))))-).)).... ( -21.80, z-score = -0.46, R) >droAna3.scaffold_12929 845466 109 + 3277472 CCAGCCUGCUGCCUCAUAUCAGAAUCAGAAUCAGUUGCUACCGCACCAAGCUUUUCAUUGCCCGCCAACUCA----UCCCAGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCG ........(((..((......))..))).....((((..((((((....((((((((..((((.........----......))))..))))....)))))))))-)..)))). ( -23.76, z-score = 0.77, R) >dp4.chrXL_group1e 7626323 92 - 12523060 ---------------CCGCCAUCAGCUGAA-CUCUGGCUACCGCACCAAGCUUUCCAUUGCCC-CCAAUCAU----AGCCUGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCG ---------------.(((....((((...-....))))((((((....((((((((..((((-(.......----.....)))))..))))....)))))))))-)....))) ( -28.10, z-score = -1.26, R) >droPer1.super_13 1344841 92 + 2293547 ---------------CCGCCAUCAGCUGAA-CUCUGGCUACCGCACCAAGCUUUCCAUUGCCC-CCAAUCAU----AGCCUGGGGCUUUGGAUAAAAAGUUGUGG-UUAUAGCG ---------------.(((....((((...-....))))((((((....((((((((..((((-(.......----.....)))))..))))....)))))))))-)....))) ( -28.10, z-score = -1.26, R) >droWil1.scaffold_181096 3326913 85 + 12416693 -------------------------GAACAU-UG-UUUUACCACACCAAGCUUUUCAUUGCC-UUCAAACAAUUAAUGCCAGAGGCUUUGGAUAAAAAGUUGUGG-UUAUAUAG -------------------------......-((-(...((((((....((((((((..(((-((....((.....))...)))))..))))....)))))))))-)...))). ( -17.40, z-score = -1.20, R) >droVir3.scaffold_12932 1537746 85 - 2102469 --------------------CGUCAGAGCGUCUA-UGAUACCACACCAAGCUUUUCAUUGCC--GCAAACA------GCCAGAGGCUCUGGAUAAAAAGUUGUGGCUUAUAUCA --------------------...((((((.(((.-..............((........)).--((.....------)).))).))))))((((..((((....)))).)))). ( -19.20, z-score = -0.28, R) >droMoj3.scaffold_6473 13871643 75 + 16943266 ----------------------------CGCCUG-UCUUACCACACCAAGCUUUCCAUUGCC--GCAAACU------GCCCGAGGCACUGGAUAAAAAG-UGUGG-UUAUAGUA ----------------------------...(((-(...(((((((.......((((.((((--((.....------))....)))).))))......)-)))))-).)))).. ( -23.72, z-score = -2.57, R) >droGri2.scaffold_15203 6328957 80 + 11997470 ------------------------CGUCAGUCCAGUGCUACCACACCAAGCUUUUCAUUGCC--GCAAAACA-------CAGAGGCUCUGGAUAAAAAGUUGUGG-UUACAGCA ------------------------...........((((((((((....((((((((..(((--.(......-------..).)))..))))....)))))))))-)...)))) ( -18.00, z-score = -0.06, R) >consensus ____________________AC_AAAUCAGUCUAGUCUUACCGCACCAAGCUUUCCAUUGCCC_CCAAACUA_____GCCAGGGGCUCUGGAUAAAAAGUUGUGG_UUAUAGCG ........................................(((((........((((..(((.....................)))..))))........)))))......... (-14.14 = -13.68 + -0.46)
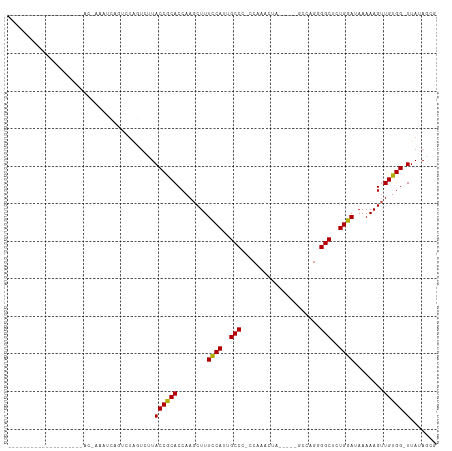
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:48 2011