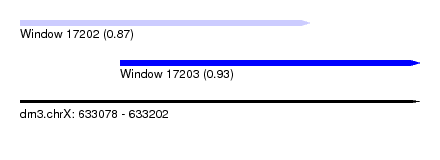
| Sequence ID | dm3.chrX |
|---|---|
| Location | 633,078 – 633,202 |
| Length | 124 |
| Max. P | 0.933053 |

| Location | 633,078 – 633,168 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 88.64 |
| Shannon entropy | 0.18238 |
| G+C content | 0.44326 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 633078 90 + 22422827 GCUGCCUUCUGCUUUCCGCCCACUCCCUGCCCCCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUUGUACAUGCAAUAUUUUGGGU ((........))........................((((..(((((((.(((((((...........))))))).)))))))..)))). ( -15.40, z-score = -0.76, R) >droYak2.chrX 558089 86 + 21770863 GCCGAACUCUGCCCUCUGUA----CCCUGAAUUCUGUCCAUUGUGCUGUUUGUGUAAUUAAAGUUGAUUUGCACAUGCAAUAUUUUGGGU ..........((((...(((----(..((.........))..))))(((.(((((((...........))))))).))).......)))) ( -16.30, z-score = -0.59, R) >droSec1.super_10 482162 89 + 3036183 GCUGCCUUCUGCUUUCCGCC-ACUGCCUGCCCUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUUGCACAUGCAAUAUUUUGGGU ((.((.....((.....)).-...))..))......((((..(((((((.(((((((...........))))))).)))))))..)))). ( -21.90, z-score = -1.92, R) >droSim1.chrX 519654 89 + 17042790 GCUGCCUUCUGCUUUCCGCC-ACUGCCUGCCCUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUUGCACAUGCAAUAUUUUGGGU ((.((.....((.....)).-...))..))......((((..(((((((.(((((((...........))))))).)))))))..)))). ( -21.90, z-score = -1.92, R) >consensus GCUGCCUUCUGCUUUCCGCC_ACUCCCUGCCCUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUUGCACAUGCAAUAUUUUGGGU ((........))........................((((..(((((((.(((((((...........))))))).)))))))..)))). (-18.04 = -18.10 + 0.06)
| Location | 633,109 – 633,202 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Shannon entropy | 0.45751 |
| G+C content | 0.40834 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
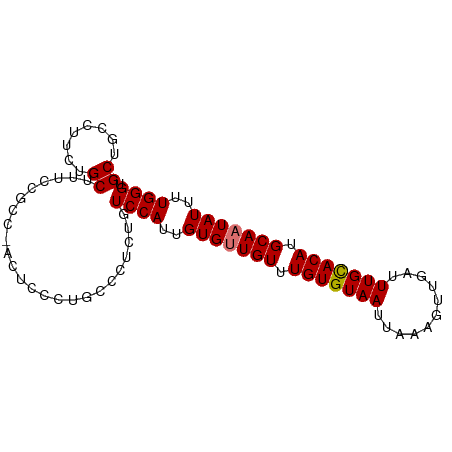
>dm3.chrX 633109 93 + 22422827 CCCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUU------GUACAUGCAAUAUUUUGGGUCCCGCGCCACCAAGUGUGUUGUGCGUAUUUUGUG .....((((..(((((((.(((((((...........))------))))).)))))))..))))...(((((.(((....).)).)))))......... ( -21.80, z-score = -1.65, R) >droEre2.scaffold_4644 597410 91 + 2521924 -UCUGUCCAUUGUGCUGUUUGUGUAAUUAAAGUUGAUUU------GCACAUGCAAUAUUUUGGGAGCGGCGCU-CGCAGAGUGUUGUGCGUACUUUGUG -......((..((((....(((((((...........))------))))).(((((((((((.((((...)))-).)))))))))))..))))..)).. ( -29.30, z-score = -2.00, R) >droSec1.super_10 482192 93 + 3036183 CUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUU------GCACAUGCAAUAUUUUGGGUCCCGCGCCACCAAGUGUGUUGUGCGUAUUUUGUG .....((((..(((((((.(((((((...........))------))))).)))))))..))))...(((((.(((....).)).)))))......... ( -26.70, z-score = -2.83, R) >droSim1.chrX 519684 93 + 17042790 CUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUU------GCACAUGCAAUAUUUUGGGUCCCGCGCCACCAAGUGUGUUGUGCGUAUUUUGUG .....((((..(((((((.(((((((...........))------))))).)))))))..))))...(((((.(((....).)).)))))......... ( -26.70, z-score = -2.83, R) >droWil1.scaffold_180777 2699226 93 + 4753960 UUUUAUGCAUUUU----UGUGUGUAAUUAAAGUUGAUUUUGGCUAACACGAAGGGAGCCUUUAAGCGAAUGGCCCAAUGUAUGGUAAAAAGACUUUA-- ..((((((((...----.)))))))).(((((((..(((((.((((((.....((.(((...........)))))..))).)))))))).)))))))-- ( -19.20, z-score = -0.50, R) >consensus CUCUGUCCAUUGUGUUGUUUGUGUAAUUAAAGUUGAUUU______GCACAUGCAAUAUUUUGGGUCCCGCGCCACCAAGUGUGUUGUGCGUAUUUUGUG .....((((..(((((((.(((((....(((.....)))......))))).)))))))..))))....((((.((.......)).)))).......... (-13.70 = -14.58 + 0.88)

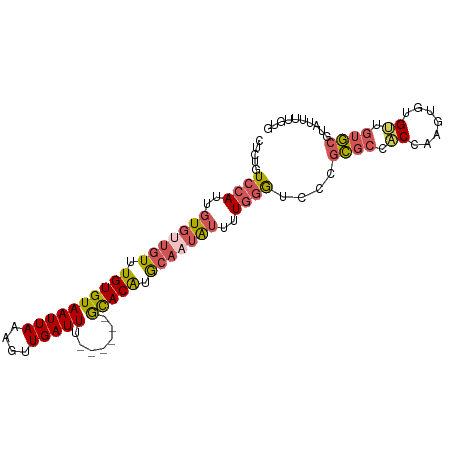
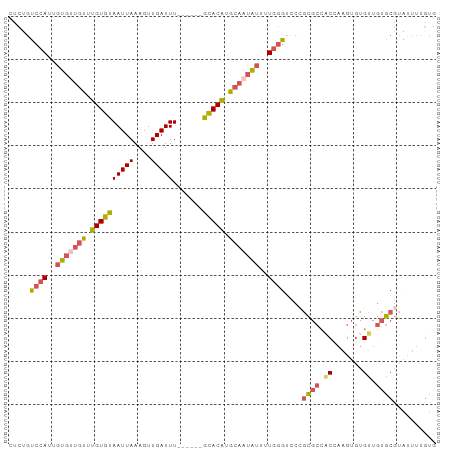
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:39 2011