
| Sequence ID | dm3.chrX |
|---|---|
| Location | 626,661 – 626,728 |
| Length | 67 |
| Max. P | 0.526605 |

| Location | 626,661 – 626,728 |
|---|---|
| Length | 67 |
| Sequences | 9 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 77.44 |
| Shannon entropy | 0.44599 |
| G+C content | 0.34696 |
| Mean single sequence MFE | -10.97 |
| Consensus MFE | -5.34 |
| Energy contribution | -6.16 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 626661 67 + 22422827 ------GGGGAAUUUUGCAUUUUAACGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA-AGUCGCACAAGU----- ------......(((((((((..............))))))))).....((((........-))))........----- ( -10.94, z-score = -1.45, R) >droEre2.scaffold_4644 590940 67 + 2521924 ------GGAGAAUUUAGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA-AGUCGCAGCCGU----- ------((..(((((.(((((...(((.....))))))))..)))))..((((........-))))....))..----- ( -11.70, z-score = -1.38, R) >droYak2.chrX 551413 67 + 21770863 ------GCAGAAUUUAGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA-AGUCGCACCAGU----- ------((..(((((.(((((...(((.....))))))))..)))))..((((........-))))))......----- ( -12.50, z-score = -2.36, R) >droSec1.super_10 475870 67 + 3036183 ------GGGAAAUUUUGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA-AGUAGCACAAGU----- ------......(((((((((...(((.....))))))))))))......(((........-))).........----- ( -8.80, z-score = -0.67, R) >droSim1.chrX 513263 67 + 17042790 ------GGGAAAUUUUGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA-AGUCGCACAAGU----- ------......(((((((((...(((.....)))))))))))).....((((........-))))........----- ( -12.50, z-score = -1.98, R) >dp4.chrXL_group1e 121575 78 + 12523060 GCUGUGGGACAAUUUUGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACACAAACACACACACACAU- ..(((.((....(((((((((...(((.....)))))))))))).......)).))).....................- ( -9.60, z-score = -1.03, R) >droPer1.super_60 189770 76 - 401158 GCUGUAGGACAAUUUUGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACACAAACACACACACAU--- ..(((((.....(((((((((...(((.....)))))))))))).......)).)))...................--- ( -8.20, z-score = -0.85, R) >droAna3.scaffold_13117 659523 63 + 5790199 ----------AACUCUGCAUUUUAAUGAACUCCAUAAUGCGAAAAUUAAGACUGACACACG-UGUCACACCCGG----- ----------.....((((((...(((.....)))))))))...........(((((....-))))).......----- ( -9.00, z-score = -1.13, R) >droVir3.scaffold_12970 10573958 72 - 11907090 ------GGACAAUUUCGCAUAUUAAUGAAUUUAAUAUUGCGAAAAUAAAGACAGCUAAAGG-ACACAUGCUGAACGAGU ------......(((((((((((((.....)))))).))))))).......((((......-......))))....... ( -15.50, z-score = -3.61, R) >consensus ______GGACAAUUUUGCAUUUUAAUGAAUUUCAUAAUGCGAAAAUUAAGACUGACACACA_AGUCGCACAAGU_____ ............(((((((((...(((.....))))))))))))................................... ( -5.34 = -6.16 + 0.82)

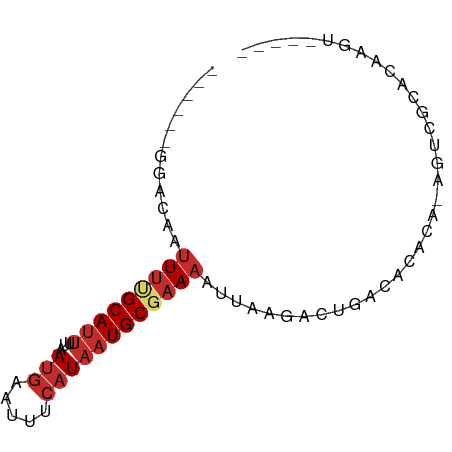
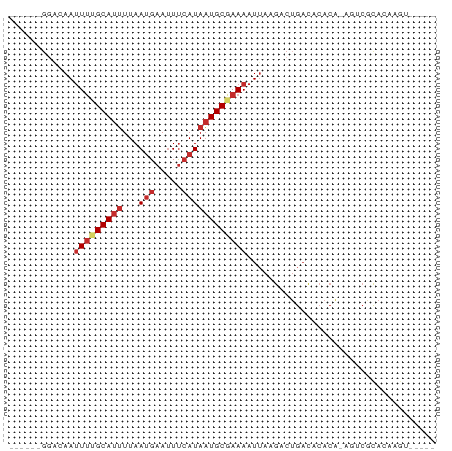
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:37 2011