| Sequence ID | dm3.chrX |
|---|---|
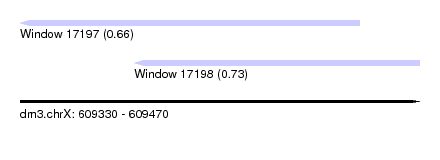
| Location | 609,330 – 609,470 |
| Length | 140 |
| Max. P | 0.728478 |

| Location | 609,330 – 609,449 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Shannon entropy | 0.10032 |
| G+C content | 0.59558 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -29.81 |
| Energy contribution | -29.37 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656325 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 609330 119 - 22422827 GCUUAGCUGCCUCGCAGCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGCAGCUUCAGUAGUGGCUCCUCCUCAGCUACUUUCCCCUACCUUCCGACCCCUUCUCCAUCCUGU ....((((((...))))))(((..((((((.....)))))).(((((((...)))))))......(((((((.......)))))))..............)))................ ( -36.00, z-score = -2.57, R) >droSim1.chrX 494685 116 - 17042790 GCUUAGCUGCCUCG--GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGUAGCUUCAGUAGUGGCUCCUCCUCGGCC-CCUUUCCCUACUCCCCAGCCCCUUCUCCAUCCUGU ((......))...(--((((....((((......))))(((((((((((...))))))).......(.((.....)).)))))-...............)))))............... ( -31.60, z-score = -0.95, R) >droSec1.super_10 457692 116 - 3036183 GCUUAGCUGCCUCG--GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGUAGCUUCAGUAGUGGCUCCUCCUCGGCC-CCUUUCCCUACUCCCCAGCCCCUUCUCCAUCCUGU ((......))...(--((((....((((......))))(((((((((((...))))))).......(.((.....)).)))))-...............)))))............... ( -31.60, z-score = -0.95, R) >consensus GCUUAGCUGCCUCG__GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGUAGCUUCAGUAGUGGCUCCUCCUCGGCC_CCUUUCCCUACUCCCCAGCCCCUUCUCCAUCCUGU (((.((..(((.(...((((....((((((.....)))))).(((((((...)))))))..)))).).))).....)).)))..................................... (-29.81 = -29.37 + -0.44)



| Location | 609,370 – 609,470 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Shannon entropy | 0.17226 |
| G+C content | 0.60026 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.73 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 609370 100 - 22422827 GCACACGGCCACUCUGCUCGAGCUUAGCUGCCUCGCAGCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGCAGCUUCAGUAGUGGCUCCUCCUC ((.(((((((.(((.....)))....((((....((((((....)))))).....))))))))(((((((...))))))).......)))))........ ( -42.80, z-score = -2.46, R) >droSim1.chrX 494724 98 - 17042790 GCACACGGCCACUCUGCUCGAGCUUAGCUGCCUCG--GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGUAGCUUCAGUAGUGGCUCCUCCUC ......((((((((((...(((((.((((((..((--(((.....((((......)))))))))..)))))).....)))))))).)))))))....... ( -40.40, z-score = -2.33, R) >droSec1.super_10 457731 98 - 3036183 GCACACGGCCACUCUGCUCGAGCUUAGCUGCCUCG--GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGUAGCUUCAGUAGUGGCUCCUCCUC ......((((((((((...(((((.((((((..((--(((.....((((......)))))))))..)))))).....)))))))).)))))))....... ( -40.40, z-score = -2.33, R) >droEre2.scaffold_4644 572447 92 - 2521924 -CACACGGCCAUUCUGCUCGAGCUUAGCCACCUCG--GCUGUCAAGUUGCUUUAUCAGCGGCCGCUGCAGCUCCUGCAGCUUCUGUAGCUUCGGU----- -.(((.((((...(((...((((.(((((.....)--)))).......))))...))).))))(((((((...)))))))...))).((....))----- ( -31.50, z-score = -0.62, R) >consensus GCACACGGCCACUCUGCUCGAGCUUAGCUGCCUCG__GCUGUCAAGCUGCUUUAUCAGCGGCCGCUGCAGCUUCUGCAGCUUCAGUAGUGGCUCCUCCUC .......((((((((((..(((((((((.(((.....(((....))).(((.....)))))).)))).)))))..)))).......))))))........ (-30.55 = -30.73 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:35 2011