| Sequence ID | dm3.chrX |
|---|---|
| Location | 582,859 – 582,960 |
| Length | 101 |
| Max. P | 0.658072 |

| Location | 582,859 – 582,960 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Shannon entropy | 0.41473 |
| G+C content | 0.43925 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 582859 101 - 22422827 ------------CCCCAUGCUGA-AAACCCCAGUGCA----AAUGUGAGGUUAUGGGCGUAAGCCCCUCGCGAGCUGCGAAUCGAAAAUUUGUUGAUUUUGUAUUUAAUAUUUAGUUU ------------......(((((-(......((((((----(((((((((.....(((....))))))))))..(.((((((.....)))))).)..)))))))).....)))))).. ( -25.20, z-score = -1.82, R) >droSim1.chrX 468168 101 - 17042790 ------------CCCCAUGCUGG-AAACCCCAGUGCA----AAUGUGAGGUUAUGGGCGUAAGCCCCUCGCGAGCUGCGAAUCUGAAAUUUGUUGAUUUUGUAUUUAAUAUUUAGUUU ------------......(((((-.....)))))..(----((((..((((((.((((....)))).((((.....)))).............))))))..)))))............ ( -27.20, z-score = -1.93, R) >droSec1.super_10 430248 101 - 3036183 ------------UCCCAUGCUGG-AAACCCCAGUGCA----AAUGUGAGGUUAUGGGCGUAAGCCCCUCGCGAGCUGCGAAUCUGAAAUUUGUUGAUUUUGUAUUUAAUAUUUAGUUU ------------......(((((-.....)))))..(----((((..((((((.((((....)))).((((.....)))).............))))))..)))))............ ( -27.20, z-score = -1.85, R) >droEre2.scaffold_4644 546269 82 - 2521924 --------------------------------GUGCA----AAUGUGAGGUUAUGGGCGUGAGCCCCUCGCGAGCUGCGAAUCGGAAAAUUGUUGACUCUGUAUUUAAUAUUUAGUUU --------------------------------(((((----...((((((.....(((....)))))))))((((.((((.........)))).).)))))))).............. ( -21.70, z-score = -1.22, R) >droAna3.scaffold_13117 618910 117 - 5790199 AGCCCGCGAGUACGAAACACAAACAAAGCCCAGUGCGAGGCAGUGCGAGGUUAUGUGUGUAAGCCCC-CGCAGAACCCAAAUCGCAAAUUUGUUGAUUCUGUAUUUAAUAUUUGGCUU ((((...(((((((((.((((((....(((........)))..(((((((((.((((.(......).-)))).))))....)))))..)))).)).))).)))))).......)))). ( -27.40, z-score = -0.17, R) >consensus ____________CCCCAUGCUGA_AAACCCCAGUGCA____AAUGUGAGGUUAUGGGCGUAAGCCCCUCGCGAGCUGCGAAUCGGAAAUUUGUUGAUUUUGUAUUUAAUAUUUAGUUU ...............................((((((......(((((((.....(((....))))))))))....((((((.....))))))......))))))............. (-16.06 = -16.26 + 0.20)

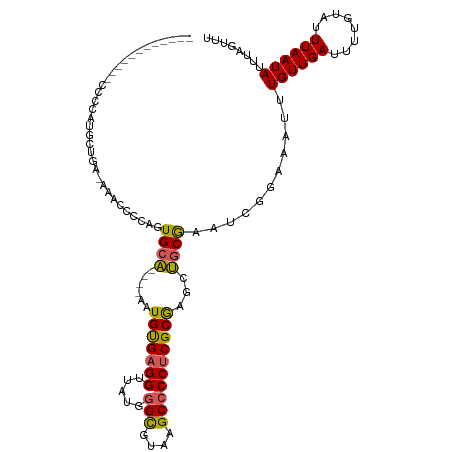

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:25 2011