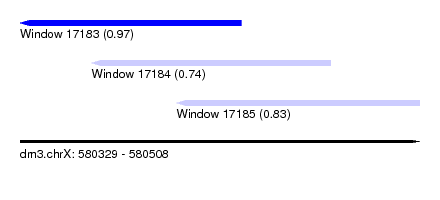
| Sequence ID | dm3.chrX |
|---|---|
| Location | 580,329 – 580,508 |
| Length | 179 |
| Max. P | 0.973892 |

| Location | 580,329 – 580,428 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.31 |
| Shannon entropy | 0.19730 |
| G+C content | 0.34751 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 580329 99 - 22422827 UCUGAGUGCCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGAAAAAGAAAGCGCACUAAUUGUGUUUUGUAUUACGAUGAAA-------UAAUACUCAAACCCAAAAA ..(((((((((..(((.......)))..))))...((((.((.(.((((((((.....)))))))).).)).))))....-------....))))).......... ( -21.10, z-score = -2.29, R) >droSim1.chrX 465641 99 - 17042790 UCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGAAAGCGCGCUAAUUGUGUUUUGCAUUACGAUGAAA-------UAAUACUUAAAACCGAAAA ..((((((...(((((((((((......))))..)))))))....((((((((.....))))))))..............-------...)))))).......... ( -21.20, z-score = -0.77, R) >droSec1.super_10 427681 106 - 3036183 UCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGAAAGCGCGCUAAUUGUGUUUUGUAUUACGAUGAAAUAUAAUAUAAUACUUAAAACCAAAAA ..((((((((.(((((((((((......))))..)))))))...(....)))....(((((((((..(......)..)))))))))....)))))).......... ( -22.70, z-score = -1.60, R) >droYak2.chrX 505583 98 - 21770863 UUUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGAAAGCGCACUAAUUGUGUUUUGUAUUACAAUAAAA-------UAACACUCAAAACUAAAA- ((((((((...(((((((((((......))))..)))))))..(.((((((((.....)))))))).)............-------...)))))))).......- ( -28.40, z-score = -3.95, R) >droEre2.scaffold_4644 543693 99 - 2521924 UUUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAUGAAAGCCCACUAAUUGUGUUUUGUAUUACAAUUAAA-------UAAUGCUCAAAACUGUGGA ((((((((((.(((((((((((......))))..))))))).......)))...((((((((........))))))))..-------....)))))))........ ( -25.10, z-score = -2.05, R) >consensus UCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGAAAGCGCACUAAUUGUGUUUUGUAUUACGAUGAAA_______UAAUACUCAAAACCAAAAA ..((((((...(((((((((((......))))..)))))))....((((((((.....))))))))........................)))))).......... (-21.66 = -21.26 + -0.40)

| Location | 580,361 – 580,468 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.42045 |
| G+C content | 0.43994 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -17.85 |
| Energy contribution | -19.75 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 580361 107 - 22422827 ACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGCCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGAAAAAGA--AAGCGCACUAAUUGUGUUUUG (((((.(((((((..((.((........))))..))))))).)))))((((..(((.......)))..)))).............(--(((((((.....)))))))). ( -30.10, z-score = -1.89, R) >droSim1.chrX 465673 107 - 17042790 ACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGA--AAGCGCGCUAAUUGUGUUUUG (((((.(((((((..((.((........))))..))))))).)))))....(((((((((((......))))..)))))))....(--(((((((.....)))))))). ( -33.70, z-score = -2.15, R) >droSec1.super_10 427720 107 - 3036183 ACUCAAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGA--AAGCGCGCUAAUUGUGUUUUG (((((.(((((((..((.((........))))..))))))).)))))....(((((((((((......))))..)))))))....(--(((((((.....)))))))). ( -34.40, z-score = -2.50, R) >droYak2.chrX 505614 107 - 21770863 ACUCGAAAUGGGCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGA--AAGCGCACUAAUUGUGUUUUG (((((((((((.(...((((........))))..).)))))))))))....(((((((((((......))))..)))))))....(--(((((((.....)))))))). ( -32.30, z-score = -2.07, R) >droEre2.scaffold_4644 543725 107 - 2521924 ACUCAAAACGGCCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAUGA--AAGCCCACUAAUUGUGUUUUG ((((((((..(((...((((........))))..)))..))))))))(((.(((((((((((......))))..))))))).....--..)))(((.....)))..... ( -30.30, z-score = -1.91, R) >droVir3.scaffold_12970 10476957 104 + 11907090 GAUGCUGUCUGCUCAGUGCAUUUAACUCAGAAU--GCCACUCAAAGUAAC-UUCACUCGCGGA--AAGCGCUCAAGUCAGCCGCGGUUAAAUUCAAAAGGUGUGCUAGG ((((((.(((((..(((((((((......))))--).))))...(((...-...))).)))))--.)))).)).....((((((..((.......))..))).)))... ( -23.00, z-score = 0.88, R) >consensus ACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGCCGUUUGAUUGGCUAAGUUGGAAAAGA__AAGCGCACUAAUUGUGUUUUG (((((.(((((((.......((((....))))..))))))).)))))....((((((.((((......))))...)))))).......(((((((.....))))))).. (-17.85 = -19.75 + 1.90)

| Location | 580,399 – 580,508 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.44597 |
| G+C content | 0.48803 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -16.04 |
| Energy contribution | -17.74 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 580399 109 - 22422827 GCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAAUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGCCAUCCAAUUGC------CGUUUGAUU----- ((((...((((.((......))))))...)).)).....((((((.(((((((..((.((........))))..))))))).))))))............------.........----- ( -25.40, z-score = -0.93, R) >droSim1.chrX 465711 109 - 17042790 GCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAGUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGC------CGUUUGAUU----- .....(((((((.(((.............)))))))))).(((((.(((((((..((.((........))))..))))))).)))))((((......)))------)........----- ( -32.02, z-score = -2.09, R) >droSec1.super_10 427758 109 - 3036183 GCCUCACUGUGCAGUUUCCUCUCACAAUCAGCGCACAGUCACUCAAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGC------CGUUUGAUU----- .....(((((((.(((.............)))))))))).(((((.(((((((..((.((........))))..))))))).)))))((((......)))------)........----- ( -32.72, z-score = -2.77, R) >droYak2.chrX 505652 109 - 21770863 GCACCACUGUGCAGUUUCCUCUCAAAUCCAGCGCAAAGUCACUCGAAAUGGGCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGCAUCCAAUUGC------CGUUUGAUU----- ((((....))))............((((.(((((((.((((((((((((((.(...((((........))))..).))))))))))))))......))).------)))).))))----- ( -30.70, z-score = -1.69, R) >droEre2.scaffold_4644 543763 109 - 2521924 GCCUCACUGUGCGAUUUCCUCUCGAAGCCAGCGCACAGUCACUCAAAACGGCCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGCAUCCAAUUGC------CGUUUGAUU----- .....((((((((.((((.....))))....)))))))).((((((((..(((...((((........))))..)))..))))))))((((......)))------)........----- ( -35.20, z-score = -3.16, R) >droVir3.scaffold_12970 10476984 117 + 11907090 GUAUCACAGACAACAAUUCUCAUACCACUGUCGCGCAACAGAUGCUGUCUGCUCAGUGCAUUUAACUCAGAAU--GCCACUCAAAGUAAC-UUCACUCGCGGAAAGCGCUCAAGUCAGCC ........(((................((((......))))..(((.(((((..(((((((((......))))--).))))...(((...-...))).))))).)))......))).... ( -23.80, z-score = -0.05, R) >consensus GCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAGUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGCAUCCAAUUGC______CGUUUGAUU_____ ........((((..................))))...((((((((.(((((((.......((((....))))..))))))).)))))))).............................. (-16.04 = -17.74 + 1.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:24 2011