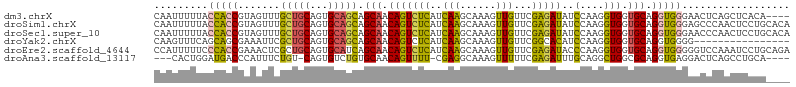
| Sequence ID | dm3.chrX |
|---|---|
| Location | 561,579 – 561,681 |
| Length | 102 |
| Max. P | 0.847438 |

| Location | 561,579 – 561,681 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.50944 |
| G+C content | 0.50644 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -11.97 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
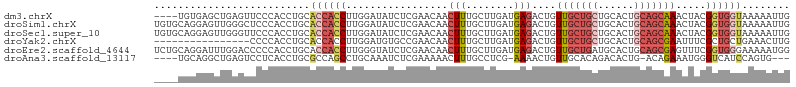
>dm3.chrX 561579 102 + 22422827 ----UGUGAGCUGAGUUCCCACCUGCACCACCUUGGAUAUCUCGAACAACUUUGCUUGAUGAGACUGUUGCUGCUGCACUGCAGCAAACUACGGUGGUAAAAAUUG ----((.((((...)))).)).....((((((.(((...(((((..(((......))).)))))...(((((((......))))))).))).))))))........ ( -28.30, z-score = -1.77, R) >droSim1.chrX 444963 106 + 17042790 UGUGCAGGAGUUGGGCUCCCACCUGCACCACCUUGGAUAUCUCGAACAACUUUGCUUGAUGAGACUGUUGCUGCUGCACUGCAGCAAACUACGGUGGUAAAAAUUG ...(((((...(((....))))))))((((((.(((...(((((..(((......))).)))))...(((((((......))))))).))).))))))........ ( -38.50, z-score = -3.30, R) >droSec1.super_10 407382 106 + 3036183 UGUGCAGGAGUUGGGUUCCCACCUGCACCACCUUGGAUAUCUCGAACAACUUUGCUUGAUGAGACUGUUGCUGCUGCACUGCAGCAAACUACGGUGGUAAAAAUUG ...(((((...(((....))))))))((((((.(((...(((((..(((......))).)))))...(((((((......))))))).))).))))))........ ( -38.50, z-score = -3.32, R) >droYak2.chrX 481291 90 + 21770863 ----------------CCCCACCUGCACCACCUUGGAUGUGCCGAACAACUUUGCUUGAUGAGACUGUUGCUGCUGCACUGCAGCGAAUUUCGCUGCUGAAACUUG ----------------...........((.....))..((((.(..((((....(((...)))...))))...).)))).((((((.....))))))......... ( -21.00, z-score = -1.68, R) >droEre2.scaffold_4644 523168 106 + 2521924 UCUGCAGGAUUUGGACCCCCACCUGCACCACCUUGGGUAUCUCGAACAACUUUGCUUGAUGAGACUGUUGCUGAUGCACUGCAGCGAGUUUCGGUGGGAAAAAUGG ..((((((...(((....)))))))))((((((..((((.............))))..).(((((((((((.........))))).)))))))))))......... ( -36.12, z-score = -2.22, R) >droAna3.scaffold_13117 587785 97 + 5790199 ----UGCAGGCUGAGUCCUCACCUGCGCCAGCCUGCAAAUCUCGAAAAACUUUGCCUCG-AAAACUGUUGCACAGACACUG-ACAGAAAUGGGUCAUCCAGUG--- ----(((((((((.((.(......).)))))))))))....((((...........)))-)...(((.....))).((((.-.((....))((....))))))--- ( -27.10, z-score = -1.15, R) >consensus ____CAGGAGUUGGGUCCCCACCUGCACCACCUUGGAUAUCUCGAACAACUUUGCUUGAUGAGACUGUUGCUGCUGCACUGCAGCAAACUACGGUGGUAAAAAUUG ..........................((((((.................(((........)))....(((((((......))))))).....))))))........ (-11.97 = -13.30 + 1.33)
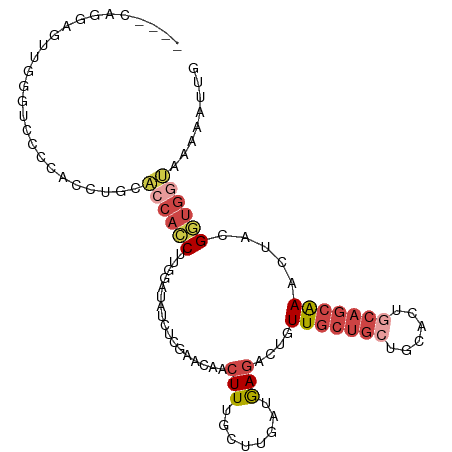
| Location | 561,579 – 561,681 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.50944 |
| G+C content | 0.50644 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.05 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 561579 102 - 22422827 CAAUUUUUACCACCGUAGUUUGCUGCAGUGCAGCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGAGAUAUCCAAGGUGGUGCAGGUGGGAACUCAGCUCACA---- .......(((((((...(((.(((((......)))))))).(((((..(((......)))...)))))......)))))))...(((((.......))))).---- ( -32.00, z-score = -1.98, R) >droSim1.chrX 444963 106 - 17042790 CAAUUUUUACCACCGUAGUUUGCUGCAGUGCAGCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGAGAUAUCCAAGGUGGUGCAGGUGGGAGCCCAACUCCUGCACA ........((((((...(((.(((((......)))))))).(((((..(((......)))...)))))......))))))((((((((....)))...)))))... ( -37.80, z-score = -2.85, R) >droSec1.super_10 407382 106 - 3036183 CAAUUUUUACCACCGUAGUUUGCUGCAGUGCAGCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGAGAUAUCCAAGGUGGUGCAGGUGGGAACCCAACUCCUGCACA ........((((((...(((.(((((......)))))))).(((((..(((......)))...)))))......))))))((((((((....)))...)))))... ( -37.80, z-score = -3.12, R) >droYak2.chrX 481291 90 - 21770863 CAAGUUUCAGCAGCGAAAUUCGCUGCAGUGCAGCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGGCACAUCCAAGGUGGUGCAGGUGGGG---------------- .........((((((.....))))))..(((....))).....((((((.((((....))))..((((..(....)..)))).)))))).---------------- ( -26.60, z-score = -1.21, R) >droEre2.scaffold_4644 523168 106 - 2521924 CCAUUUUUCCCACCGAAACUCGCUGCAGUGCAUCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGAGAUACCCAAGGUGGUGCAGGUGGGGGUCCAAAUCCUGCAGA .........(((((.......((((........))))....(((((..(((......)))...)))))......)))))(((((((((....)))...)))))).. ( -29.40, z-score = -0.27, R) >droAna3.scaffold_13117 587785 97 - 5790199 ---CACUGGAUGACCCAUUUCUGU-CAGUGUCUGUGCAACAGUUUU-CGAGGCAAAGUUUUUCGAGAUUUGCAGGCUGGCGCAGGUGAGGACUCAGCCUGCA---- ---...(((.....))).....((-((((.....(((((....(((-(((((.......)))))))).))))).))))))((((((((....))).))))).---- ( -28.90, z-score = 0.39, R) >consensus CAAUUUUUACCACCGUAGUUCGCUGCAGUGCAGCAGCAACAGUCUCAUCAAGCAAAGUUGUUCGAGAUAUCCAAGGUGGUGCAGGUGGGAACCCAACUCCUG____ .........(((((.......(((((...))))).(((.(((((((..(((......)))...)))))..(....))).))).))))).................. (-15.54 = -15.05 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:19 2011