| Sequence ID | dm3.chrX |
|---|---|
| Location | 555,795 – 555,909 |
| Length | 114 |
| Max. P | 0.596044 |

| Location | 555,795 – 555,909 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Shannon entropy | 0.44219 |
| G+C content | 0.41576 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 555795 114 + 22422827 ----AUAUAUAAUAUCCGUUUCACUGAAUAUUGUGCUGCAAUUAGCAGCAG-AGUUCAGCUGACAGCUGUUGCAACGAAUUCUCAAUCGAGUAAUAGCAGAAUAGCAGAAUCGAAUCGG ----...........(((.(((..(((((..(.((((((.....)))))).-))))))((((...((((((((..(((........))).))))))))....))))......))).))) ( -32.60, z-score = -2.39, R) >droYak2.chrX 476579 114 + 21770863 ----ACAUAUAAUAUCCGUUUCACUGCAUAUUGUGCUGCAAUUAGCAGCGG-AGUUCAGCUGACAGCUGUUGCAUCGAAUUCCCAAUCGAGUAAUAGCAGAAUAGCACAAUCGAGUCGG ----...........(((.(((.......(((.((((((.....)))))).-)))...((((...((((((((.((((........))))))))))))....))))......))).))) ( -33.10, z-score = -2.23, R) >droEre2.scaffold_4644 512006 106 + 2521924 ----ACACGUAAUAUCCGUUUCACUGAAUAUUGUGCUGCAAUUAGCAGCAG-GGUUCAGCUGCCAGCUGUUGCACCGAAUUUUCAAUCGAGUAAUAGCACAAUAGCAGAAG-------- ----...................(((..((((((((((......(((((((-(((......)))..)))))))(((((........))).))..)))))))))).)))...-------- ( -30.00, z-score = -2.03, R) >droSec1.super_10 401414 114 + 3036183 ----AUAUAUAAUAUCCGUUUCACUGAAUAUUGUGCUGCAAUUAGCAGCAG-AGUUCAGCUGACAGCUGUUGCACGGAAUUCUCAAUCGAGUAAUAGCAGAAUAGCAGAAUCGAAUCGG ----...........(((.(((..(((((..(.((((((.....)))))).-))))))((((...((((((((.(((.........))).))))))))....))))......))).))) ( -32.50, z-score = -2.19, R) >droSim1.chrX 438692 118 + 17042790 AUAUACAUAUAAUAUCCGUUUCACUGAAUAUUGUGCUGCAAUUAGCAGCAG-AGUUCAGCUGACAGCUGUUGCACGGAAUUCCCAAUCGAGUAAUAGCAGAAUAGCAAAAUCGAAUCGG ...............(((.(((..(((((..(.((((((.....)))))).-))))))((((...((((((((.(((.........))).))))))))....))))......))).))) ( -32.50, z-score = -2.66, R) >dp4.chrXL_group1e 8015 107 + 12523060 ----CUCUAUAAUUUCCGUUCGAUUGCAUAUUUUGUUGCAAUUAGCAGCAGAAGUUCAGCUGACGGCUUUUGCAUGGAAUUCCCAAUUUGGAAU--AUCGAAUAUCAAAUGCG------ ----..........((((((.(((((((........)))))))))).(((((((((........)))))))))..)))(((((......)))))--.................------ ( -25.40, z-score = -1.11, R) >droPer1.super_60 74850 107 - 401158 ----CUCUAUAAUUUCCGUUCGAUUGCAUAUUUUGUUGCAAUUAGCAGCAGAAGUUCAGCUGAUGGCUUUUGCAUGGAAUUCCCAAUUUGGAAU--AUCGAAUAUCAAAUGCG------ ----..........((((((.(((((((........)))))))))).((((((((.((.....))))))))))..)))(((((......)))))--.................------ ( -25.70, z-score = -1.08, R) >droAna3.scaffold_13117 582827 94 + 5790199 --------------UCCGGACGACUGAAUAUUUUGCUGCAAUUAGCAGCA--AGUCCAGCCGACAGCUGUUGCAUCGAAUGG-CAAUCGAGGAU--GCGGAAUAUC-GAGUCGA----- --------------......(((((..((((((((((((.....))))).--.((((...(((..((((((......)))))-)..))).))))--..))))))).-.))))).----- ( -30.40, z-score = -1.39, R) >consensus ____AUAUAUAAUAUCCGUUUCACUGAAUAUUGUGCUGCAAUUAGCAGCAG_AGUUCAGCUGACAGCUGUUGCAUCGAAUUCCCAAUCGAGUAAUAGCAGAAUAGCAGAAUCGA_____ .................(((...(((.......((((((.....))))))...((.((((.....))))..))........................)))...)))............. (-13.57 = -13.70 + 0.13)

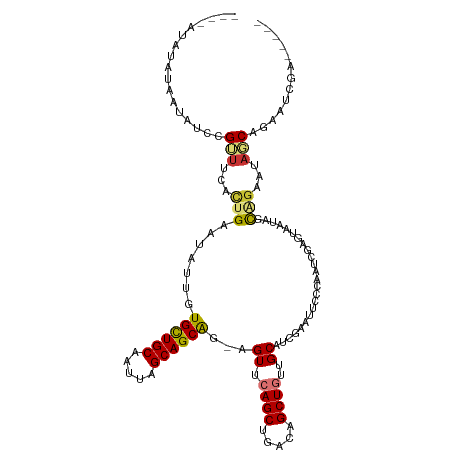

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:17 2011