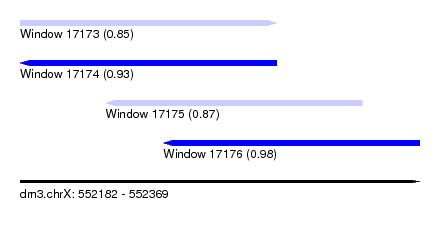
| Sequence ID | dm3.chrX |
|---|---|
| Location | 552,182 – 552,369 |
| Length | 187 |
| Max. P | 0.975451 |

| Location | 552,182 – 552,302 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.36430 |
| G+C content | 0.42191 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -21.34 |
| Energy contribution | -22.65 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847005 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 552182 120 + 22422827 ACUGUAGCUGCCACUUUGCCACUUAGUGUGCAAUUGAAAUGCGCGCCAACGCAUGUUCUCAUAUGCGGAGCGUUUAGCAGCCCUUAAAAAGCUUUUCAUGUUGCUUUUAAAAAGCUACGA ..(((((((((((..(((((((...))).)))).))...(((((((...(((((((....)))))))..))))...))))).....((((((..........))))))....))))))). ( -34.20, z-score = -1.06, R) >droSim1.chrX 434820 110 + 17042790 --------ACUGUGGCUGCCACUUGGUA--CAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGUUUCGCAGCACUUAAAAAGCUUUUCAUUUUGCUUUUAAAAAGCUACGA --------..((((((((((....))).--.....(((((((((.....(((((........))))))))))))))..........((((((..........))))))....))))))). ( -30.90, z-score = -1.30, R) >droSec1.super_10 397560 98 + 3036183 --------------------ACUUGGUA--CAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGCUUCGCAGCACUUAAAAAGCUUUUCAUUUUGCUUUUUAAAAGCUACGA --------------------........--..........(((((((...((((........))))))))))).(((.(((..(((((((((..........)))))))))..))).))) ( -30.00, z-score = -2.81, R) >droEre2.scaffold_4644 508104 95 + 2521924 -------------------CAUCUACAG--CAAUUGAAAAGCGCGCCAACGCAUUGU-UAAUAU---GUGCGUGUUGCAGCACUUGAAAAGCUUUUCAUGUUGCUUUUCAAAAGCUACGA -------------------.......((--(..(((((((((((((..((((((...-......---))))))...)).))((.(((((....))))).)).)))))))))..))).... ( -28.10, z-score = -1.58, R) >consensus ___________________CACUUAGUA__CAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGUUUCGCAGCACUUAAAAAGCUUUUCAUGUUGCUUUUAAAAAGCUACGA ........................................(((((((...((((........))))))))))).(((.(((..(((((((((..........)))))))))..))).))) (-21.34 = -22.65 + 1.31)

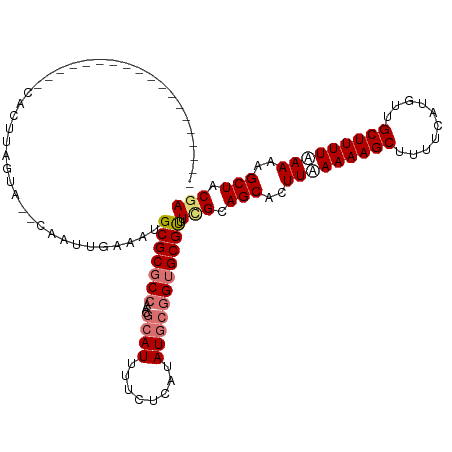
| Location | 552,182 – 552,302 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.36430 |
| G+C content | 0.42191 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -22.08 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930909 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 552182 120 - 22422827 UCGUAGCUUUUUAAAAGCAACAUGAAAAGCUUUUUAAGGGCUGCUAAACGCUCCGCAUAUGAGAACAUGCGUUGGCGCGCAUUUCAAUUGCACACUAAGUGGCAAAGUGGCAGCUACAGU ..(((((((((.((((((..........)))))).))))))))).....(((((((......(((.((((((....)))))))))..((((.(((...))))))).)))).)))...... ( -39.50, z-score = -2.03, R) >droSim1.chrX 434820 110 - 17042790 UCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUG--UACCAAGUGGCAGCCACAGU-------- (((((((.(((.((((((..........)))))).))).)))))))...((.((((.((..(..((((((((....))))))))...)..--))....))))..))......-------- ( -30.50, z-score = -0.75, R) >droSec1.super_10 397560 98 - 3036183 UCGUAGCUUUUAAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAGCGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUG--UACCAAGU-------------------- (((((((.((((((((((..........)))))))))).))))))).((((..(((((........)))))...))))(((.......))--).......-------------------- ( -34.00, z-score = -3.34, R) >droEre2.scaffold_4644 508104 95 - 2521924 UCGUAGCUUUUGAAAAGCAACAUGAAAAGCUUUUCAAGUGCUGCAACACGCAC---AUAUUA-ACAAUGCGUUGGCGCGCUUUUCAAUUG--CUGUAGAUG------------------- (..((((..(((((((((....(((((....))))).((((((....(((((.---......-....))))))))))))))))))))..)--)))..)...------------------- ( -29.40, z-score = -2.02, R) >consensus UCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUG__CACCAAGUG___________________ (((((((.((((((((((..........)))))))))).)))))))..(((..(((((........)))))...)))........................................... (-22.08 = -23.70 + 1.62)

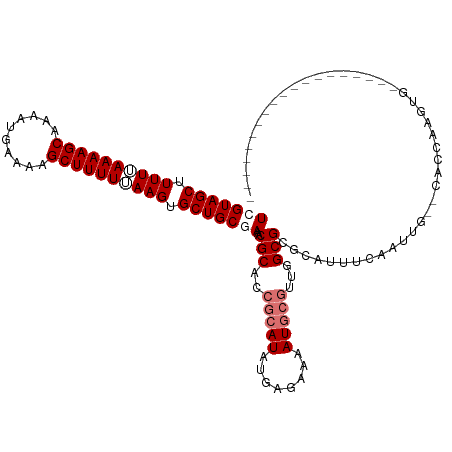

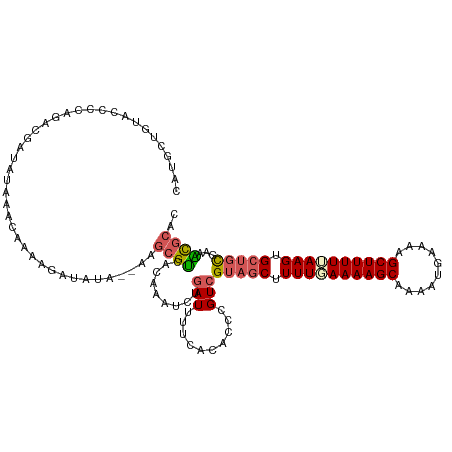
| Location | 552,222 – 552,342 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.34990 |
| G+C content | 0.41819 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -20.54 |
| Energy contribution | -22.74 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 552222 120 - 22422827 AAAGAUAUAUAAAGCGUCACAAAUCUAAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAACAUGAAAAGCUUUUUAAGGGCUGCUAAACGCUCCGCAUAUGAGAACAUGCGUUGGCGCGC .............((((((...................(((.(((((((((.((((((..........)))))).)))))))))...)))...(((((........))))).)))))).. ( -30.40, z-score = -1.55, R) >droSim1.chrX 434850 116 - 17042790 AAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGC ........----.((((((.....................(((((((.(((.((((((..........)))))).))).))))))).......(((((........))))).)))))).. ( -30.30, z-score = -1.23, R) >droSec1.super_10 397578 116 - 3036183 AAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUAAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAGCGCACCGCAUAUGAGAAAAUGCGUUGGCGCGC ........----.((((((......))))...........(((((((.((((((((((..........)))))))))).))))))).((((..(((((........)))))...)))))) ( -35.10, z-score = -2.42, R) >droYak2.chrX 472638 105 - 21770863 AUAAAUAA----AGCGCCAAAAGUCAGAUUCUUACACCCGUCGCAGCUUUUGAAAAGCAAAAUGAAAAGCUUUUCAAGUG----AAAUCGCGU----UGGUACGAAGUG-GUAGCCAC-- ........----.((((((.(((.......))).....((((.((((..(((((((((..........)))))))))(((----....)))))----))).)))...))-)).))...-- ( -22.50, z-score = -0.75, R) >droEre2.scaffold_4644 508123 112 - 2521924 AAAAAUUA----AGCGUCAAAAAUCGGAUUCCCACACCCGUCGUAGCUUUUGAAAAGCAACAUGAAAAGCUUUUCAAGUGCUGCAACACGCA---CAUAUUA-ACAAUGCGUUGGCGCGC ........----.((((((......((....))......((.(((((.((((((((((..........)))))))))).))))).))(((((---.......-....))))))))))).. ( -34.20, z-score = -3.48, R) >consensus AAAGAUAU____AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUGAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCAAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGC .............(((((........................(((((.((((((((((..........)))))))))).)))))..((((((...............))))))))))).. (-20.54 = -22.74 + 2.20)

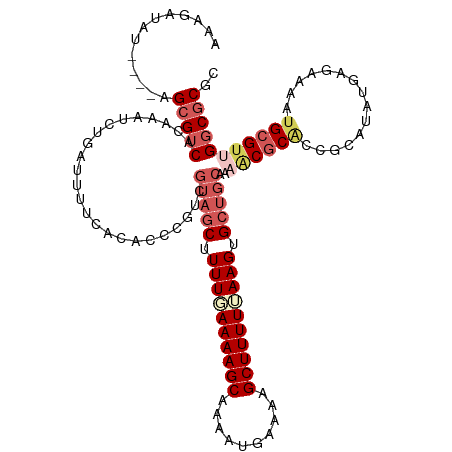

| Location | 552,249 – 552,369 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.32 |
| Shannon entropy | 0.43997 |
| G+C content | 0.39629 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 552249 120 - 22422827 CAUGCUGUACCCCAGACGAUAUAAAAAAAAGAUAUAUAAAGCGUCACAAAUCUAAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAACAUGAAAAGCUUUUUAAGGGCUGCUAAACGCUC ...((.........((((.((((...........))))...))))........................(((((((((.((((((..........)))))).))))))))).....)).. ( -21.50, z-score = -1.24, R) >droSim1.chrX 434877 116 - 17042790 CAUGCUGUACCCCAGACGAUAUAAACAAAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCAC ..(((.........((((.((((.........)))----).))))......................(((((((.(((.((((((..........)))))).))).)))))))...))). ( -24.20, z-score = -2.02, R) >droSec1.super_10 397605 116 - 3036183 CAUGCUGUACCCCAGACGAUAUAAACAAAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUAAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAGCGCAC ...(((........((((.((((.........)))----).))))......................(((((((.((((((((((..........)))))))))).)))))))))).... ( -28.60, z-score = -3.16, R) >droYak2.chrX 472658 101 - 21770863 ------------CAUGCUGU-UCCCCAAAAUAAAUA--AAGCGCCAAAAGUCAGAUUCUUACACCCGUCGCAGCUUUUGAAAAGCAAAAUGAAAAGCUUUUCAAGUGAAAUCGCGU---- ------------...(((((-..(........(((.--..((.......))...))).........)..)))))..(((((((((..........)))))))))(((....)))..---- ( -16.03, z-score = -0.37, R) >droEre2.scaffold_4644 508146 108 - 2521924 ------------CAUGCGAUGUACAUACACCAAAAAUUAAGCGUCAAAAAUCGGAUUCCCACACCCGUCGUAGCUUUUGAAAAGCAACAUGAAAAGCUUUUCAAGUGCUGCAACACGCAC ------------..((((.(((.............................(((..........)))..(((((.((((((((((..........)))))))))).))))).))))))). ( -27.30, z-score = -3.01, R) >consensus CAUGCUGUACCCCAGACGAUAUAAACAAAAGAUAUA__AAGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUGAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCAC ........................................((((.........(((..........)))(((((.((((((((((..........)))))))))).)))))...)))).. (-15.84 = -16.56 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:17 2011