
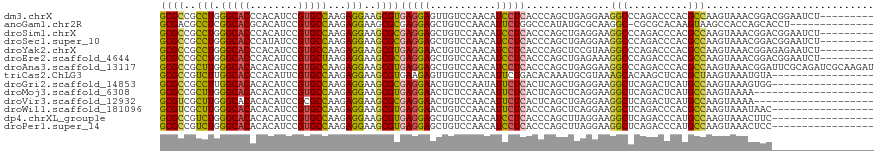
| Sequence ID | dm3.chrX |
|---|---|
| Location | 522,481 – 522,591 |
| Length | 110 |
| Max. P | 0.597987 |

| Location | 522,481 – 522,591 |
|---|---|
| Length | 110 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Shannon entropy | 0.40460 |
| G+C content | 0.58685 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 522481 110 - 22422827 GCGCCGCCUGGGCACCCACAUCCGUGCCAAGAGGAAGCGUGAGGAGUUGUCCAACAUCCUCACCCAGCUGAGGAAGGCCCAGACCCACGCCAAGUAAACGGACGGAAUCU--------- ((((..(((.(((((........)))))...)))..)))).(((..((((((....((((((......)))))).(((..........)))........))))))..)))--------- ( -37.10, z-score = -1.02, R) >anoGam1.chr2R 33109793 104 + 62725911 GCGACGCCUCGGCACGCACAUCCGUGCCAAGAGGAAGCGCGAGGAGCUGUCCAACAUUCUGGCCCAUAUGCGCAAGGC-CGCGCACAAGUAAGCCACCAGCACCU-------------- (((...((((((((((......))))))..))))...))).(((.((((..........((((.....(((((.....-.))))).......)))).)))).)))-------------- ( -39.40, z-score = -1.75, R) >droSim1.chrX 405766 110 - 17042790 GCGCCGCCUGGGCACCCACAUCCGUGCCAAGAGGAAGCGCGAGGAGCUGUCCAACAUCCUCACCCAGCUGAGGAAGGCCCAGACCCACGCCAAGUAAACGGACGGAAUCU--------- ((((..(((.(((((........)))))...)))..)))).(((..((((((....((((((......)))))).(((..........)))........))))))..)))--------- ( -41.90, z-score = -2.24, R) >droSec1.super_10 368793 110 - 3036183 GCGCCGCCUGGGCACCCAUAUCCGUGCCAAGAGGAAGCGCGAGGAGCUGUCCAACAUCCUCACCCAGCUGAGGAAGGCCCAGACCCACGCCAAGUAAACGGACGGAAUCU--------- ((((..(((.(((((........)))))...)))..)))).(((..((((((....((((((......)))))).(((..........)))........))))))..)))--------- ( -41.90, z-score = -2.35, R) >droYak2.chrX 443533 110 - 21770863 GCGCCGCCUGGGCACCCACAUCCGUGCCAAGAGGAAGCGUGAGGAACUGUCCAACAUCCUCACCCAGCUCCGUAAGGCCCAGACCCACGCCAAGUAAACGGAGAGAAUCU--------- ..((..(((.(((((........)))))...)))..))(((((((..((.....)))))))))....((((((..(((..........)))......)))))).......--------- ( -34.10, z-score = -1.45, R) >droEre2.scaffold_4644 477056 110 - 2521924 GCGCCGCCUGGGCACCCACAUCCGUGCUAAGAGGAAGCGCGAGGAGCUGUCCAACAUCCUCACCCAGCUGAGAAAGGCCCAGACCCACGCCAAGUAAACGGACGGAAUCU--------- ((((..(((.(((((........)))))...)))..)))).(((..((((((......((((......))))...(((..........)))........))))))..)))--------- ( -35.10, z-score = -0.84, R) >droAna3.scaffold_13117 3017308 119 + 5790199 GCGCCGCUUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAGCUGUCCAACAUCCUCACCCAGCUGAGGAAGGCCCAGACCCACGCCAAGUAAACGGAUUCGCAGAUCGCAAGAU (((((((((((((((........)))))........(((((.((..(((..(....((((((......))))))..)..))).)))))))))))....)))...)))..(((....))) ( -41.10, z-score = -1.81, R) >triCas2.ChLG3 6313964 102 - 32080666 GCGCCGUCUUGGCACCCACAUUCGUGCCAAGAGGAAGCGUGAAGAGUUGUCCAACAUUCUGACACAAAUGCGUAAAGCACAAGCUCACGCUAAGUAAAUGUA----------------- ((..(.(((((((((........))))))))).)..))(((.(((((.(.....))))))..)))....((((..(((....))).))))............----------------- ( -31.60, z-score = -2.22, R) >droGri2.scaffold_14853 7614728 102 + 10151454 GCGCCGCCUUGGCACACACAUCCGUGCCAAGAGGAAGCGCGAGGAACUGUCCAAUAUUCUCACUCAGCUGAGGAAGGCUCAGACUCAUGCCAAGUAAAGUGG----------------- ((((..(((((((((........))))))..)))..))))(((...(((.((....((((((......)))))).))..))).)))................----------------- ( -32.10, z-score = -0.73, R) >droMoj3.scaffold_6308 3024917 99 + 3356042 GCGCCGCUUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAACUCUCCAACAUUCUCACUCAGCUCAGGAAGGCUCAGACUCAUGCCAAGUAAAA-------------------- .....((((((((((........)))))........(((((((((....)))..........((.((((......)))).))..)))))))))))....-------------------- ( -28.10, z-score = -0.46, R) >droVir3.scaffold_12932 240819 99 - 2102469 GCGUCGCUUGGGCACACACAUCCGCGCCAAGAGGAAGCGCGAGGAACUGUCCAACAUUCUCACUCAGCUGAGGAAGGCUCAGACUCAUGCCAAGUAAAA-------------------- .....((((((.((.........((((.........))))(((...(((.((....((((((......)))))).))..))).))).))))))))....-------------------- ( -25.90, z-score = 0.61, R) >droWil1.scaffold_181096 1249170 102 - 12416693 GCGUCGCUUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAACUGUCCAACAUUCUCACCCAGCUCAGGAAGGCUCAGACCCACGCCAAGUAAAUAAC----------------- .....((((((((((........)))))........(((((.((..(((..(....((((...........)))).)..))).)))))))))))).......----------------- ( -26.40, z-score = -0.07, R) >dp4.chrXL_group1e 1611838 102 + 12523060 GCGCCGUCUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAGCUGUCCAACAUCCUCACCCAGCUUAGGAAGGCUCAGACCCAUGCCAAGUAAACUUC----------------- ((..(.(((.(((((........))))).))).)..))(((((((..((.....)))))))))((......))..(((..........)))...........----------------- ( -30.80, z-score = -0.62, R) >droPer1.super_14 1816133 102 + 2168203 GCGCCGUCUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAGCUGUCCAACAUCCUCACCCAGCUUAGGAAGGCUCAGACCCAUGCCAAGUAAACUCC----------------- ((..(.(((.(((((........))))).))).)..))(((((((..((.....)))))))))((......))..(((..........)))...........----------------- ( -30.80, z-score = -0.58, R) >consensus GCGCCGCCUGGGCACACACAUCCGUGCCAAGAGGAAGCGUGAGGAGCUGUCCAACAUCCUCACCCAGCUGAGGAAGGCCCAGACCCACGCCAAGUAAACGGA_________________ ((((..(((.(((((........)))))...)))..))))(((((...........)))))..............(((..........)))............................ (-23.88 = -23.08 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:13 2011