
| Sequence ID | dm3.chrX |
|---|---|
| Location | 494,445 – 494,565 |
| Length | 120 |
| Max. P | 0.874212 |

| Location | 494,445 – 494,565 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.47 |
| Shannon entropy | 0.53766 |
| G+C content | 0.46716 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.874212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 494445 120 + 22422827 AUAUCUGCAGCUACUAUUAAUACCUUUCACUGAAACAAAGGGUAUCAAAUAGUCGCGAGCGCUGUCUAUUGCGUUCUCUCUUAUGUUUUCAGAGGAGGGGAAACUGUUUCACAGCGAAUU ......((.((.((((((.((((((((..........))))))))..)))))).))..))(((((..((.(..(((((((((.((....)).)))))))))..).))...)))))..... ( -34.00, z-score = -2.50, R) >droEre2.scaffold_4644 446624 112 + 2521924 ------UCAGGUGCAUCGUGCUUCCGUUGGUUGAAUAGCGGGUAUCUAAUAGUCGAGAGCGCCUCCCACAGCGUCCUCUUUUAUGAUUUCGUAAGAGGGGAA--UGCUUCGCAGCGAACU ------((((((((.(((.((((((((((......)))))))........))))))..)))))).....((((((((((((((((....))))))))))).)--)))).......))... ( -38.30, z-score = -1.64, R) >droYak2.chrX 414381 95 + 21770863 ------UCAGCUCCAUCGUACUUCCUUUGGCUGAAUAGGGGGUAUCCAAUAGUCG------------------UCCUCUCGCAUGUUUUCGUAAAAGGGGAAA-UGCUUCGCAGCGAAUG ------...(((.(...(((((((((.(......).))))))))).....(((..------------------((((((...(((....)))...))))))..-.)))..).)))..... ( -21.00, z-score = 0.59, R) >consensus ______UCAGCUACAUCGUACUUCCUUUGGUUGAAUAGAGGGUAUCAAAUAGUCG_GAGCGC___C_A__GCGUCCUCUCUUAUGUUUUCGUAAGAGGGGAAA_UGCUUCGCAGCGAAUU ......................(((((((......)))))))..........((...................((((((((((((....))))))))))))....(((....)))))... (-18.73 = -18.63 + -0.09)

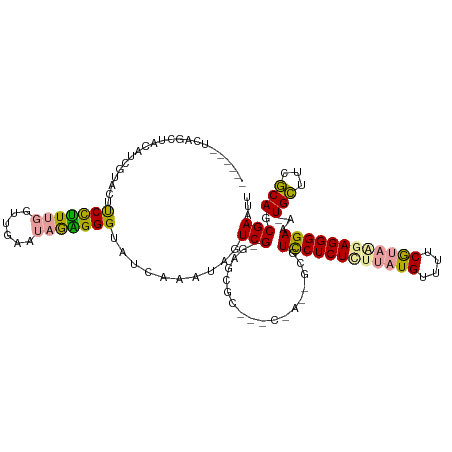
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:08 2011