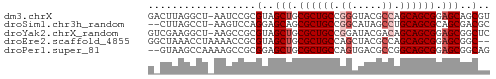
| Sequence ID | dm3.chrX |
|---|---|
| Location | 445,764 – 445,820 |
| Length | 56 |
| Max. P | 0.946925 |

| Location | 445,764 – 445,820 |
|---|---|
| Length | 56 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.48655 |
| G+C content | 0.67392 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -19.84 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
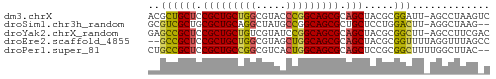
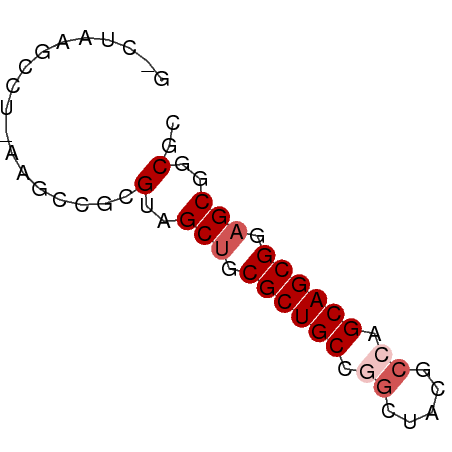
>dm3.chrX 445764 56 + 22422827 ACGCUGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUU-AGCCUAAGUC ..((((.(((((((((((.....))))))))((......))))).)-)))....... ( -25.40, z-score = -1.71, R) >droSim1.chr3h_random 862076 54 + 1452968 GCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGACUU-AGGCUAAG-- .....((.(((((((.(((...))).))))))).)).((((....)-))).....-- ( -23.20, z-score = -0.31, R) >droYak2.chrX_random 1263066 56 - 1802292 GAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUU-AGCCUUCGAC (((((((...((((((((((....)))))..)))))...)))))))-.......... ( -25.70, z-score = -2.09, R) >droEre2.scaffold_4855 234991 55 - 242605 --GCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCC --((.(((.((((((..((...))..)))))).)))...))((((........)))) ( -24.00, z-score = -0.55, R) >droPer1.super_81 10316 55 + 277874 CUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUAC-- ((((.(((.(((((((((.....))))))))).)))...))))............-- ( -24.70, z-score = -1.27, R) >consensus GCGCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUACGCGGCUU_AGGCUUAG_C ..((((((.(((((((((.....))))))))).))).....)))............. (-19.84 = -19.68 + -0.16)
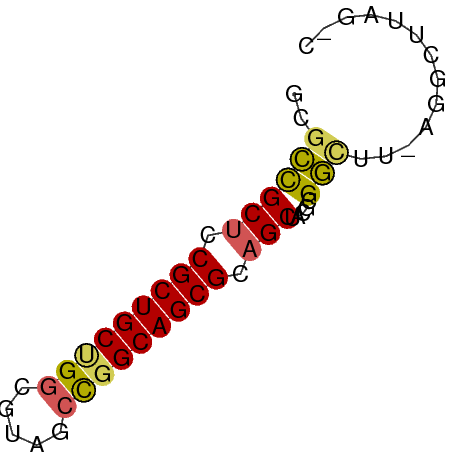
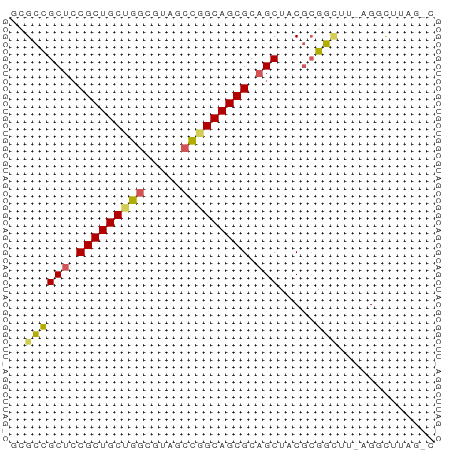
| Location | 445,764 – 445,820 |
|---|---|
| Length | 56 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.48655 |
| G+C content | 0.67392 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -17.20 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 445764 56 - 22422827 GACUUAGGCU-AAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCAGCGU ......((..-...))((((.(((.((((((.((.....)).)))))).))).)))) ( -24.80, z-score = -0.96, R) >droSim1.chr3h_random 862076 54 - 1452968 --CUUAGCCU-AAGUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGC --........-.......(..((.(((((((.(((...))).))))))).))..).. ( -23.30, z-score = -1.40, R) >droYak2.chrX_random 1263066 56 + 1802292 GUCGAAGGCU-AAGCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGCUC ..........-.((((((....((((((((.((....)).)))).)))).)))))). ( -25.30, z-score = -1.23, R) >droEre2.scaffold_4855 234991 55 + 242605 GGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGC-- ..............(((((((((((......)))))))((.....))...)))).-- ( -18.60, z-score = 0.55, R) >droPer1.super_81 10316 55 - 277874 --GUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAG --....(((.....(....).(((.(((((((.(.....).))))))).)))))).. ( -25.90, z-score = -1.25, R) >consensus G_CUAAGCCU_AAGCCGCGUAGCUGCGCUGCCGGCUACGCCAGCAGCGGAGCGGCGC ..................(..(((.((((((.((.....)).)))))).)))..).. (-17.20 = -18.00 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:03:02 2011