
| Sequence ID | dm3.chrX |
|---|---|
| Location | 432,927 – 433,032 |
| Length | 105 |
| Max. P | 0.613248 |

| Location | 432,927 – 433,032 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.58528 |
| G+C content | 0.43714 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -11.60 |
| Energy contribution | -12.55 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
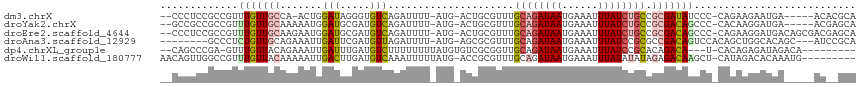
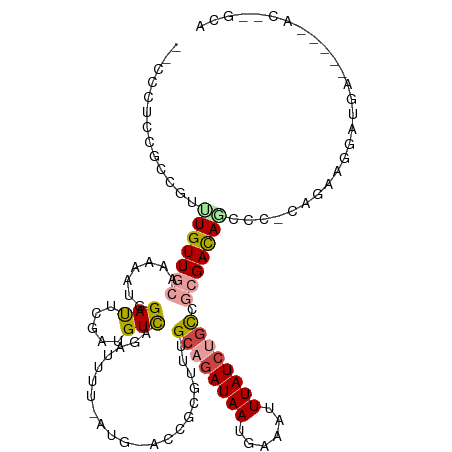
>dm3.chrX 432927 105 + 22422827 --CCCUCCGCCGUUUGUUGCCA-ACUGGAUAGGGUGUCAGAUUUU-AUG-ACUGCGUUUGCAGAUAAUGAAAUUUAUCUGCCGCGAUAUCCC-CAGAAGAAUGA-----ACACGCA --......((.(((..((.(..-.((((...(((((((((.((..-..)-)))(((...((((((((......)))))))))))))))))))-)))..)))..)-----))..)). ( -29.50, z-score = -2.21, R) >droYak2.chrX 359259 106 + 21770863 --GCCGCCGCCGUUUGUUGCAAAAAUGGAUGCGAUGUCAGAUUUU-AUG-ACUGCGUUUGCAGAUAAUGAAAUUUAUCUGCCGCGACAGCCC-CACAAGGAUGA-----ACGAGCA --......((((((((((((......(((((((..((((......-.))-)))))))))((((((((......)))))))).)))))....(-(....))..))-----))).)). ( -33.70, z-score = -2.49, R) >droEre2.scaffold_4644 384492 111 + 2521924 --CCCUCCGCCGUUUGUUGCAAGAAUGGAUGCGAUGUCAGAUUUU-AUG-ACUGCGUUUGCAGAUAAUGAAAUUUAUCUGCCGCGACAGCCC-CAGAAGGAUGACAGCGACGAGCA --.........(((((((((.....((....)).(((((..((((-..(-.((((((..((((((((......)))))))).))).))).).-..))))..)))))))))))))). ( -34.40, z-score = -2.59, R) >droAna3.scaffold_12929 325306 103 + 3277472 --------GCCCUCUGUUGCAGAAAUUGAUUCGAUGUUAGAUUUU-AUG-AGCGCGUUUGCAGAUAAUGAAAUUUAUCCGCGCCGACAGUCCACAGCUGGCACAGC---AUCCGCA --------(((..((((.((((((.(((((.....))))).))))-.((-.(((((......(((((......))))))))))))...))..))))..)))...((---....)). ( -22.10, z-score = 0.92, R) >dp4.chrXL_group1e 162714 100 + 12523060 --CAGCCCGA-GUUUGUUACAGAAAUUGAUUUGAUGUCUUUUUUUUAUGUGUCGCGGUUGCAGAUAAUGAAAUUUAUCCGCACAGACA---U-CACAGAGAUAGACA--------- --.....(((-(((.(((.....))).)))))).(((((.(((((...(((((...((.((.(((((......))))).)))).))))---)-...))))).)))))--------- ( -22.90, z-score = -1.54, R) >droWil1.scaffold_180777 2738755 105 + 4753960 AACAGUUGGCCGUUUGUUACAAAAAUUGACUUGAUGUCAAAUUUUUAUG-ACCGCGUUUGCAGAUAAUGAAAUUUAUAUAUAGAGACAAGCU-CAUAGACACAAAUG--------- ..........(((((((........(((((.....)))))...((((((-(.(..((((....((((......))))......))))..).)-)))))).)))))))--------- ( -15.70, z-score = 0.40, R) >consensus __CCCUCCGCCGUUUGUUGCAAAAAUGGAUUCGAUGUCAGAUUUU_AUG_ACCGCGUUUGCAGAUAAUGAAAUUUAUCUGCCGCGACAGCCC_CAGAAGGAUGA_____AC__GCA .............(((((((.......(((.....))).....................((((((((......)))))))).)))))))........................... (-11.60 = -12.55 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:59 2011