
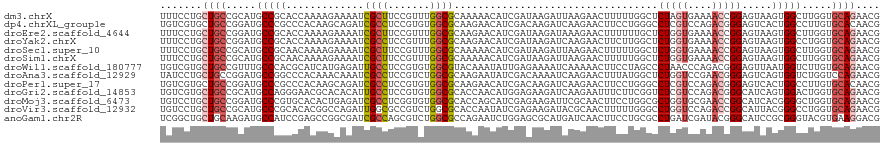
| Sequence ID | dm3.chrX |
|---|---|
| Location | 400,963 – 401,083 |
| Length | 120 |
| Max. P | 0.620687 |

| Location | 400,963 – 401,083 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.54991 |
| G+C content | 0.54679 |
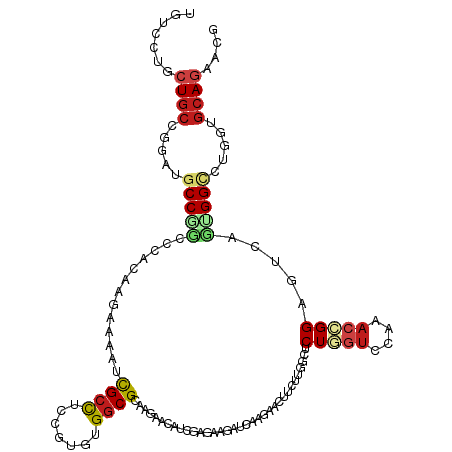
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.92 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 400963 120 + 22422827 UUUCCUGCUGCCGCAUGCCGCACCAAAAGAAAAUCGCUUCCGUUUGGCGCAAAAACAUCGAUAAGAUUAAGAACUUUUUGGCUCUAGUGAAAACCGGAGUAAGUGGCUUGGUGCAGAACG ....((((.((((...(((((.(((((((.....((((.......)))).......(((.....))).......)))))))(((..((....))..)))...))))).)))))))).... ( -34.00, z-score = -1.15, R) >dp4.chrXL_group1e 4416134 120 + 12523060 UGUCGUGCUGCCGGAUGCCCGCCCACAAGCAGAUCGCCUCCGUGUGGCGCAAGAACAUCGACAAGAUCAAGAACUUCCUGGGCCUCGUCCAGACGGGAGUCACUGGCCUUGUGCACAACG .((.((((.((((((((...((((.......((((...((.((((..(....).)))).))...)))).((......))))))..))))).(((....)))...))).....)))).)). ( -41.20, z-score = -0.32, R) >droEre2.scaffold_4644 351947 120 + 2521924 UUUCCUGCUGCCGGAUGCCGCACCAAAAGAAAAUCGCUUCCGUUUGGCGCAAGAACAUCGAUAAGAUAAAGAACUUUUUUGCUCUGGUGAAAACCGGAGUAAGUGGCUUGGUGCAGAACG ....((((.((((...(((((...(((((.....((((.......)))).......(((.....)))......)))))((((((((((....))))))))))))))).)))))))).... ( -40.30, z-score = -2.75, R) >droYak2.chrX 327080 120 + 21770863 UUUCCUGCUGCCGGAUGCCGCACCAAAAGAAAAUCGCCUCCGUUUGGCGCAAGAACAUCGAUAAGAUCAAGAACUUCUUGGCUCUGGUGAAAACCGGAGUAAGUGGCCUGGUGCAGAACG ....((((.(((((..(((((.............((((.......))))((((((..((...........))..))))))((((((((....))))))))..)))))))))))))).... ( -45.00, z-score = -3.46, R) >droSec1.super_10 256821 120 + 3036183 UUUCCUGCUGCCGCAUGCCGCAACAAAAGAAAAUCGCUUCCGUUUGGCGCAAAAACAUCGAUAAGAUUAAGAACUUUUUGGCUCUGGUGAAAACCGGAGUAAGUGGCUUGGUGCAGAACG ....((((.((((...(((((..((((((.....((((.......)))).......(((.....))).......))))))((((((((....))))))))..))))).)))))))).... ( -39.20, z-score = -2.45, R) >droSim1.chrX 294967 120 + 17042790 UUUCCUGCUGCCGCAUGCCGCAACAAAAGAAAAUCGCUUCCGUUUGGCGCAAAAACAUCGAUAAGAUUAAGAACUUUUUGGCUCUGGUGAAAACCGGAGUAAGUGGCUUGGUGCAGAACG ....((((.((((...(((((..((((((.....((((.......)))).......(((.....))).......))))))((((((((....))))))))..))))).)))))))).... ( -39.20, z-score = -2.45, R) >droWil1.scaffold_180777 4368119 120 + 4753960 UGUCGUGCUGCCGUUUGCCCACGCAUCAUGAGAUUGCCUCCGUGUGGCGUACAAAUAUUGAGAAAAUCAAAAACUUCCUAGCCCUAACCCAGACGGGAGUUAAUGGUCUUGUGCAGAACG ...(((.(((((((.((((((((......(((.....))))))).)))).)).......((((...(((..(((((((................)))))))..)))))))).)))).))) ( -29.09, z-score = -0.19, R) >droAna3.scaffold_12929 815221 120 - 3277472 UAUCCUGCUGCCGGAUGCCGGCCCACAAACAAAUCGCCUCCGUCUGGCGCAAGAAUAUCGACAAAAUCAAGAACUUUAUGGCUCUGGUCCGAACGGGAGUCAGUGGUCUGGUCCAGAACG ..((.(((.((((((((..(((.............)))..))))))))))).)).....(((.......(((.((...(((((((.((....)).)))))))..))))).)))....... ( -36.52, z-score = -0.73, R) >droPer1.super_17 1357622 120 + 1930428 UGUCGUGCUGCCGGAUGCCCGCCCACAAGCAGAUCGCCUCCGUGUGGCGCAAGAACAUCGACAAGAUCAAGAACUUCCUGGGCCUCGUCCAGACGGGAGUCACUGGCCUUGUGCACAACG .((.((((.((((((((...((((.......((((...((.((((..(....).)))).))...)))).((......))))))..))))).(((....)))...))).....)))).)). ( -41.20, z-score = -0.32, R) >droGri2.scaffold_14853 6668592 120 + 10151454 UGUCGUGCUGCCGCAUGCCAGGGAACGCACACAUUGCCUCCGUGUGGCGCACCAACAUGGAGAAGAUCAAGAAUUUCUUCGGUCUCGUCCAGACGGGCAUCAGUGGACUGGUGCAGAACG .(((((((.((((((((..(((.((........)).))).)))))))))))).....((((..((((((((.....))).)))))..)))))))..(((((((....)))))))...... ( -47.60, z-score = -1.85, R) >droMoj3.scaffold_6473 13834835 120 - 16943266 UGUCCUGCUGCCGGAUGCCCGUGCACACUGAGAUCGCCUCGGUGUGGCGCACCAGCAUCGAGAAGAUUCGCAACUUCCUGGCGCUGGUGCGAACCGGCAUCACGGGGCUGGUGCAGAACG .((.((((.(((((...(((((((((((((((.....))))))))).(((((((((..((.((((........)))).))..))))))))).........)))))).))))))))).)). ( -63.00, z-score = -4.07, R) >droVir3.scaffold_12932 1359367 120 + 2102469 UGUCCUGCUGCCGCAUGCCCGCACACGGCCAGAUUGGCGCCGUCUGGCGCACCAAUAUCGAGAAGAUACGCAACUUUUUGGGCCUGGUCCAGACCGGCAUUACGGGCCUGGUGCAGAACG .((.((((.((((...(((((......(((.....)))(((((((((...((((...((((((((........))))))))...))))))))).))))....))))).)))))))).)). ( -50.70, z-score = -2.05, R) >anoGam1.chr2R 41691016 120 - 62725911 UCGGCUGCUGCAAGAUGCCAUCCGAGCCGGCGAUCGCCAGCGUCUGGCGCCAGAAUCUGGAGCGCAUGAUCAACUUCCUGCGCCUGAUCGAUACGGGCAUCCGCGGGUACGUGAAGGACG ..(((...........))).(((..(((.((((((.((((..((((....))))..)))).(((((.((......)).)))))..)))))...).)))...((((....))))..))).. ( -44.40, z-score = 0.62, R) >consensus UGUCCUGCUGCCGGAUGCCGGCCCACAAGAAAAUCGCCUCCGUGUGGCGCAAGAACAUCGAGAAGAUCAAGAACUUCUUGGCUCUGGUCCAAACCGGAGUCAGUGGCCUGGUGCAGAACG .......((((.....(((((.............((((.......))))..................................(((((....))))).....))))).....)))).... (-16.86 = -17.92 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:51 2011