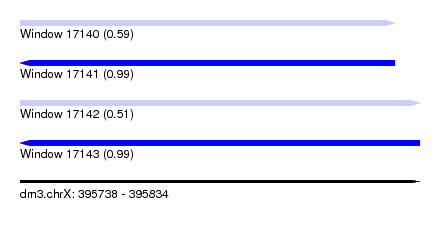
| Sequence ID | dm3.chrX |
|---|---|
| Location | 395,738 – 395,834 |
| Length | 96 |
| Max. P | 0.991090 |

| Location | 395,738 – 395,828 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.91 |
| Shannon entropy | 0.25902 |
| G+C content | 0.46502 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.56 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
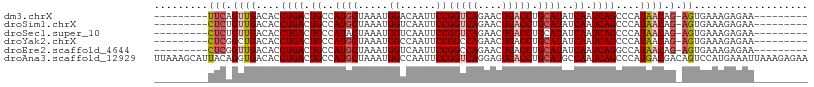
>dm3.chrX 395738 90 + 22422827 ---------UUCAGUUGACACCUGACUGCCAUGCUAAAUGGACAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG-AGUGAAAGAGAA--------- ---------(((.((((....((((.((..((((.....(((....)))(((((....))))).))))..)).))))....)))).)-))..........--------- ( -20.50, z-score = -1.28, R) >droSim1.chrX 289643 90 + 17042790 ---------CUCUGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG-AGUGAAAGAGAA--------- ---------((((((((....((((.((..((((.....((......))(((((....))))).))))..)).))))....))))))-))..........--------- ( -26.90, z-score = -3.00, R) >droSec1.super_10 251545 90 + 3036183 ---------CUCUGUUGACACCUGACUGCCAUACUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG-AGUGAAAGAGAA--------- ---------((((((((....((((..(((((.....))))).......(((((....)))))..........))))....))))))-))..........--------- ( -23.50, z-score = -2.25, R) >droYak2.chrX 321954 90 + 21770863 ---------CUCGGCUGACACCUGACUGCCAUGCUAAAUGGCCAAUUCCGGCCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG-AGUGAAAGAGAA--------- ---------(((((((((....(((.(((....(....(((((......))))).....)....)))..))).))))))(((.....-.)))...)))..--------- ( -23.80, z-score = -1.84, R) >droEre2.scaffold_4644 346780 90 + 2521924 ---------CUCGGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGCCAGAACUGACCUGCAUAUCAAUCAGGCCAUAACAG-AGUGAAAGAGAA--------- ---------(((.((((...(((((.((..((((.....(((((.(((......))).))))).))))..)).)))))...)))).)-))..........--------- ( -25.70, z-score = -1.93, R) >droAna3.scaffold_12929 809579 109 - 3277472 UUAAAGCAUUACAGGUGACACCUGACUGCCAUGCUAAAUGGCCAAUUCCGGUCAGGAGUGACCUGCAUGCCAAUCAGCCCAUGACGACAGUCCAUGAAAUUAAAGAGAA .....((((..(((((.((.((((((((((((.....))))).......))))))).)).))))).)))).........((((((....)).))))............. ( -35.11, z-score = -3.59, R) >consensus _________CUCAGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG_AGUGAAAGAGAA_________ .............((((....((((.((..((((.....((......))(((((....))))).))))..)).))))....))))........................ (-14.56 = -15.25 + 0.69)

| Location | 395,738 – 395,828 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.91 |
| Shannon entropy | 0.25902 |
| G+C content | 0.46502 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
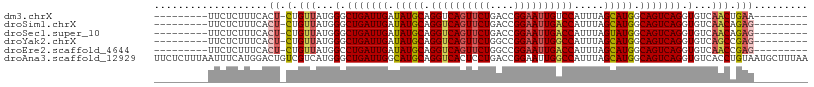
>dm3.chrX 395738 90 - 22422827 ---------UUCUCUUUCACU-CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGUCCAUUUAGCAUGGCAGUCAGGUGUCAACUGAA--------- ---------......((((((-(......)))(((((((.(((((.(((((....)))))(((....))).....))))).)))))))........))))--------- ( -27.60, z-score = -1.59, R) >droSim1.chrX 289643 90 - 17042790 ---------UUCUCUUUCACU-CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACAGAG--------- ---------..........((-(((((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...)))))))--------- ( -36.10, z-score = -4.18, R) >droSec1.super_10 251545 90 - 3036183 ---------UUCUCUUUCACU-CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGUAUGGCAGUCAGGUGUCAACAGAG--------- ---------..........((-(((((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...)))))))--------- ( -34.10, z-score = -3.72, R) >droYak2.chrX 321954 90 - 21770863 ---------UUCUCUUUCACU-CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGGCCGGAAUUGGCCAUUUAGCAUGGCAGUCAGGUGUCAGCCGAG--------- ---------..........((-(.(((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...))).)))--------- ( -30.90, z-score = -1.64, R) >droEre2.scaffold_4644 346780 90 - 2521924 ---------UUCUCUUUCACU-CUGUUAUGGCCUGAUUGAUAUGCAGGUCAGUUCUGGCCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACCGAG--------- ---------..........((-(.(((...(((((((((.(((((.((((((((((....)))))))))).....))))).)))))))))...))).)))--------- ( -36.30, z-score = -4.15, R) >droAna3.scaffold_12929 809579 109 + 3277472 UUCUCUUUAAUUUCAUGGACUGUCGUCAUGGGCUGAUUGGCAUGCAGGUCACUCCUGACCGGAAUUGGCCAUUUAGCAUGGCAGUCAGGUGUCACCUGUAAUGCUUUAA ...........((((((.((....))))))))......(((((((((((.((.((((((........(((((.....))))).)))))).)).)))))).))))).... ( -35.00, z-score = -1.57, R) >consensus _________UUCUCUUUCACU_CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACAGAG_________ ................................(((((((.(((((.((((((((((....)))))))))).....))))).)))))))..................... (-26.19 = -26.25 + 0.06)

| Location | 395,738 – 395,834 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Shannon entropy | 0.23420 |
| G+C content | 0.44863 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 395738 96 + 22422827 ---------UUCAGUUGACACCUGACUGCCAUGCUAAAUGGACAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG----------AGUGAAAGAGAAACUUUU ---------....((((....((((.((..((((.....(((....)))(((((....))))).))))..)).))))....))))((----------(((.........))))). ( -21.50, z-score = -1.46, R) >droSim1.chrX 289643 96 + 17042790 ---------CUCUGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG----------AGUGAAAGAGAAACUUUU ---------((((((((....((((.((..((((.....((......))(((((....))))).))))..)).))))....))))))----------)).(((((.....))))) ( -27.30, z-score = -3.03, R) >droSec1.super_10 251545 96 + 3036183 ---------CUCUGUUGACACCUGACUGCCAUACUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG----------AGUGAAAGAGAAACUUUU ---------((((((((....((((..(((((.....))))).......(((((....)))))..........))))....))))))----------)).(((((.....))))) ( -23.90, z-score = -2.27, R) >droYak2.chrX 321954 96 + 21770863 ---------CUCGGCUGACACCUGACUGCCAUGCUAAAUGGCCAAUUCCGGCCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG----------AGUGAAAGAGAAACUUUC ---------(((((((((....(((.(((....(....(((((......))))).....)....)))..))).))))))(((.....----------.)))...)))........ ( -23.80, z-score = -1.77, R) >droEre2.scaffold_4644 346780 96 + 2521924 ---------CUCGGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGCCAGAACUGACCUGCAUAUCAAUCAGGCCAUAACAG----------AGUGAAAGAGAAACUUUU ---------(((.((((...(((((.((..((((.....(((((.(((......))).))))).))))..)).)))))...)))).)----------)).(((((.....))))) ( -26.10, z-score = -1.96, R) >droAna3.scaffold_12929 809579 115 - 3277472 UUAAAGCAUUACAGGUGACACCUGACUGCCAUGCUAAAUGGCCAAUUCCGGUCAGGAGUGACCUGCAUGCCAAUCAGCCCAUGACGACAGUCCAUGAAAUUAAAGAGAAACUUUU .....((((..(((((.((.((((((((((((.....))))).......))))))).)).))))).)))).........((((((....)).))))................... ( -35.11, z-score = -3.36, R) >consensus _________CUCAGUUGACACCUGACUGCCAUGCUAAAUGGUCAAUUCCGGUCAGAACUGACCUGCAUAUCAAUCAGCCCAUAACAG__________AGUGAAAGAGAAACUUUU .............((((....((((.((..((((.....((......))(((((....))))).))))..)).))))....))))...............(((((.....))))) (-14.88 = -15.63 + 0.75)
| Location | 395,738 – 395,834 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.45 |
| Shannon entropy | 0.23420 |
| G+C content | 0.44863 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
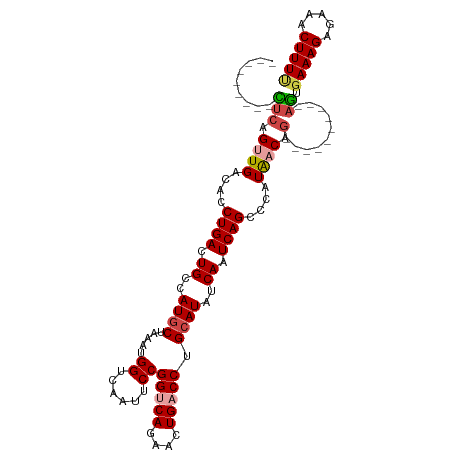
>dm3.chrX 395738 96 - 22422827 AAAAGUUUCUCUUUCACU----------CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGUCCAUUUAGCAUGGCAGUCAGGUGUCAACUGAA--------- ...((((.........((----------(......)))(((((((.(((((.(((((....)))))(((....))).....))))).))))))).....))))...--------- ( -28.00, z-score = -1.52, R) >droSim1.chrX 289643 96 - 17042790 AAAAGUUUCUCUUUCACU----------CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACAGAG--------- ................((----------(((((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...)))))))--------- ( -36.10, z-score = -3.94, R) >droSec1.super_10 251545 96 - 3036183 AAAAGUUUCUCUUUCACU----------CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGUAUGGCAGUCAGGUGUCAACAGAG--------- ................((----------(((((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...)))))))--------- ( -34.10, z-score = -3.54, R) >droYak2.chrX 321954 96 - 21770863 GAAAGUUUCUCUUUCACU----------CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGGCCGGAAUUGGCCAUUUAGCAUGGCAGUCAGGUGUCAGCCGAG--------- (((((.....))))).((----------(.(((...(.(((((((.(((((.((((((((((....)))))))))).....))))).))))))).)...))).)))--------- ( -34.80, z-score = -2.49, R) >droEre2.scaffold_4644 346780 96 - 2521924 AAAAGUUUCUCUUUCACU----------CUGUUAUGGCCUGAUUGAUAUGCAGGUCAGUUCUGGCCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACCGAG--------- ................((----------(.(((...(((((((((.(((((.((((((((((....)))))))))).....))))).)))))))))...))).)))--------- ( -36.30, z-score = -3.89, R) >droAna3.scaffold_12929 809579 115 + 3277472 AAAAGUUUCUCUUUAAUUUCAUGGACUGUCGUCAUGGGCUGAUUGGCAUGCAGGUCACUCCUGACCGGAAUUGGCCAUUUAGCAUGGCAGUCAGGUGUCACCUGUAAUGCUUUAA .(((((.................(((((((((.....(((((.((((...(.(((((....))))).).....)))).))))))))))))))(((.....))).....))))).. ( -35.60, z-score = -1.50, R) >consensus AAAAGUUUCUCUUUCACU__________CUGUUAUGGGCUGAUUGAUAUGCAGGUCAGUUCUGACCGGAAUUGACCAUUUAGCAUGGCAGUCAGGUGUCAACAGAG_________ ......................................(((((((.(((((.((((((((((....)))))))))).....))))).)))))))..................... (-26.19 = -26.25 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:50 2011