| Sequence ID | dm3.chrX |
|---|---|
| Location | 390,368 – 390,464 |
| Length | 96 |
| Max. P | 0.945078 |

| Location | 390,368 – 390,464 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.71 |
| Shannon entropy | 0.49583 |
| G+C content | 0.43928 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -18.51 |
| Energy contribution | -17.85 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
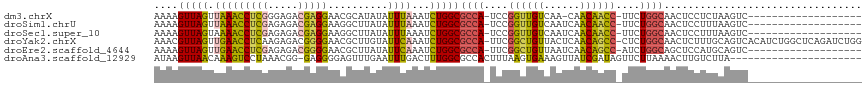
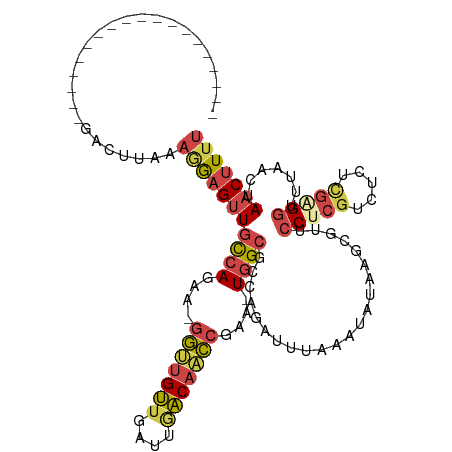
>dm3.chrX 390368 96 + 22422827 AAAAGUUAGUUAAACCUCGGGAGACGAGGAACGCAUAUAUUUAAAUCUGGCGCCA-UCCGGUUGUCAA-CAACAACC-UUCUGGCAACUCCUCUAAGUC------------------- ....(((((((((((((((.....)))))..........)))))..)))))((((-...((((((...-..))))))-...))))..............------------------- ( -23.70, z-score = -1.60, R) >droSim1.chrU 12654795 97 + 15797150 AAAAGUUAGUUAAACCUCGAGAGACGAGGAAGGCUUAUAUUUAAAUCUGGCGCCA-UCCGGUUGUCAAUCAACAACC-UUCUGGCAACUCCUUUAAGUC------------------- .((((..((((...(((((.....)))))...(((.............)))((((-...((((((......))))))-...)))))))).)))).....------------------- ( -24.22, z-score = -1.67, R) >droSec1.super_10 246094 97 + 3036183 AAAAGUUAGUAAAACCUCGAGAGACGAGGAAGGCUUAUAUUUAAAUCUGGCGCCA-UCCGGUUGUCAAUCAACAACC-UUCUGGCAACUCCUUUAAGUC------------------- ...((((.((....(((((.....))))).((..(((....)))..)).))((((-...((((((......))))))-...))))))))..........------------------- ( -23.40, z-score = -1.48, R) >droYak2.chrX 316483 116 + 21770863 AAACGUUAGUUGAACCUCAAGAGACGGGGAACGCUUGUAUUCAAAUCUGGCGCCA-UUCGGCUGUUACUCAACAGCC-CUCUGGCAACUCUUUGCAGUCACAUCUGGCUCAGAUCUGG ...((((..(((.....)))..))))...............((.(((((((((((-...(((((((....)))))))-...)))).........(((......))))).))))).)). ( -33.20, z-score = -1.11, R) >droEre2.scaffold_4644 341397 97 + 2521924 AAAAGUUAGUUGAACCUCGAGAGACGGGGAACGCUUAUAUUCAAAUCUGGCGCCA-UUCGGCUGUUAAUCAACAGCC-AUCUGGCAGCUCCAUGCAGUC------------------- ....(((((((((((((((.....)))))..........)))))..)))))((((-...(((((((....)))))))-...)))).((.....))....------------------- ( -28.90, z-score = -1.71, R) >droAna3.scaffold_12929 803864 95 - 3277472 AUAAGUUAACAAAGUCCUAAACGG-GAGGGGAGUUUGAAUUUGACUUUGGCGCCACUUUAAGUGAAAGUUAUCGAUAGUUCUUAAAACUUGUCUUA---------------------- (((((((..((((.((((...(..-..))))).))))....(((((((.((..........)).)))))))..............)))))))....---------------------- ( -19.40, z-score = -0.19, R) >consensus AAAAGUUAGUUAAACCUCGAGAGACGAGGAACGCUUAUAUUUAAAUCUGGCGCCA_UCCGGUUGUCAAUCAACAACC_UUCUGGCAACUCCUUUAAGUC___________________ ....(((((.(((((((((.....)))))..........))))...)))))((((....((((((......))))))....))))................................. (-18.51 = -17.85 + -0.66)
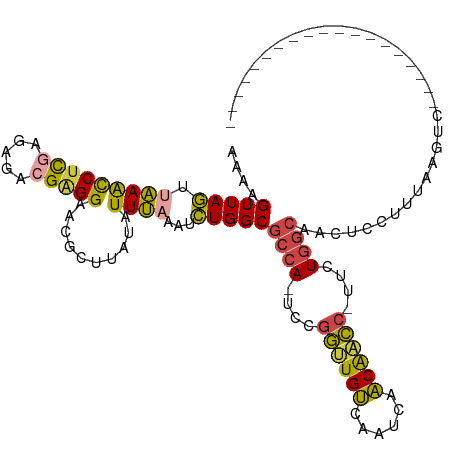
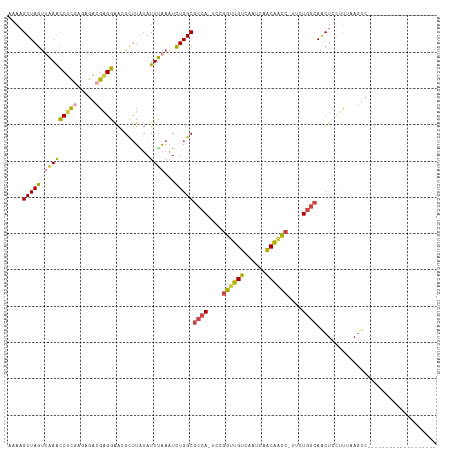
| Location | 390,368 – 390,464 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.71 |
| Shannon entropy | 0.49583 |
| G+C content | 0.43928 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 390368 96 - 22422827 -------------------GACUUAGAGGAGUUGCCAGAA-GGUUGUUG-UUGACAACCGGA-UGGCGCCAGAUUUAAAUAUAUGCGUUCCUCGUCUCCCGAGGUUUAACUAACUUUU -------------------(((...((((((..((((...-(((((((.-..)))))))...-))))((...((......))..)).)))))))))....((((((.....)))))). ( -27.10, z-score = -1.62, R) >droSim1.chrU 12654795 97 - 15797150 -------------------GACUUAAAGGAGUUGCCAGAA-GGUUGUUGAUUGACAACCGGA-UGGCGCCAGAUUUAAAUAUAAGCCUUCCUCGUCUCUCGAGGUUUAACUAACUUUU -------------------.......(((((((((((...-(((((((....)))))))...-))))......................(((((.....))))).......))))))) ( -27.10, z-score = -1.87, R) >droSec1.super_10 246094 97 - 3036183 -------------------GACUUAAAGGAGUUGCCAGAA-GGUUGUUGAUUGACAACCGGA-UGGCGCCAGAUUUAAAUAUAAGCCUUCCUCGUCUCUCGAGGUUUUACUAACUUUU -------------------.......(((((((((((...-(((((((....)))))))...-))))......................(((((.....))))).......))))))) ( -27.10, z-score = -1.84, R) >droYak2.chrX 316483 116 - 21770863 CCAGAUCUGAGCCAGAUGUGACUGCAAAGAGUUGCCAGAG-GGCUGUUGAGUAACAGCCGAA-UGGCGCCAGAUUUGAAUACAAGCGUUCCCCGUCUCUUGAGGUUCAACUAACGUUU .((((((((.((((...(..(((......)))..).....-(((((((....)))))))...-))))..)))))))).....(((((((((.((.....)).)).......))))))) ( -36.01, z-score = -1.69, R) >droEre2.scaffold_4644 341397 97 - 2521924 -------------------GACUGCAUGGAGCUGCCAGAU-GGCUGUUGAUUAACAGCCGAA-UGGCGCCAGAUUUGAAUAUAAGCGUUCCCCGUCUCUCGAGGUUCAACUAACUUUU -------------------(((.(...(((((.((((..(-(((((((....))))))))..-))))((...((......))..)))))))).)))....((((((.....)))))). ( -28.30, z-score = -1.41, R) >droAna3.scaffold_12929 803864 95 + 3277472 ----------------------UAAGACAAGUUUUAAGAACUAUCGAUAACUUUCACUUAAAGUGGCGCCAAAGUCAAAUUCAAACUCCCCUC-CCGUUUAGGACUUUGUUAACUUAU ----------------------...(((((((((((((.......((......))........((((......))))................-...)))))))).)))))....... ( -12.10, z-score = 0.22, R) >consensus ___________________GACUUAAAGGAGUUGCCAGAA_GGUUGUUGAUUGACAACCGAA_UGGCGCCAGAUUUAAAUAUAAGCGUUCCUCGUCUCUCGAGGUUUAACUAACUUUU ..........................(((((((((((....(((((((....)))))))....))))......................(((((.....))))).......))))))) (-16.02 = -16.88 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:47 2011