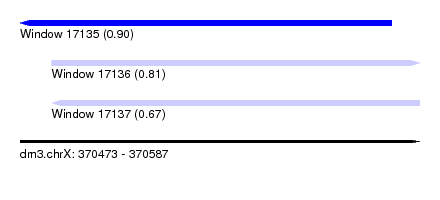
| Sequence ID | dm3.chrX |
|---|---|
| Location | 370,473 – 370,587 |
| Length | 114 |
| Max. P | 0.903749 |

| Location | 370,473 – 370,579 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.38428 |
| G+C content | 0.46973 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -17.38 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
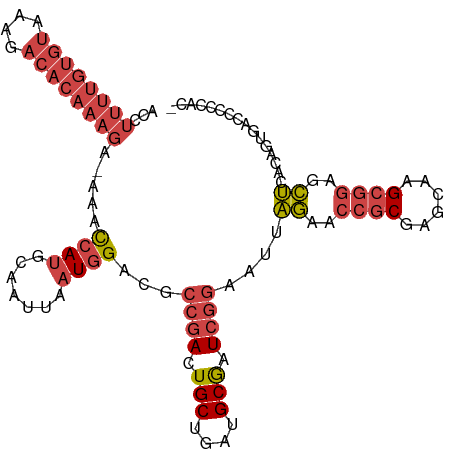
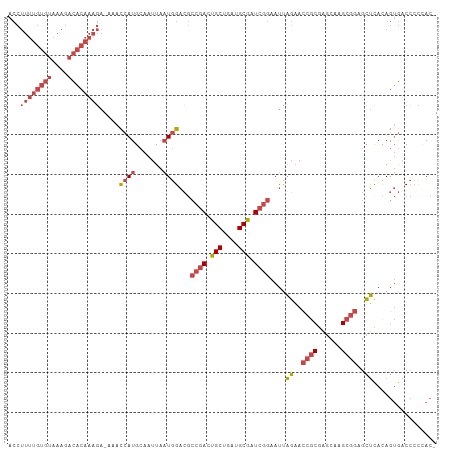
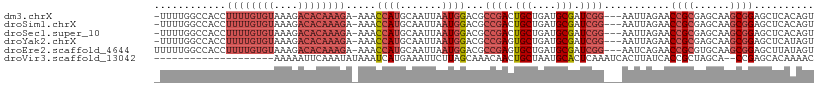
>dm3.chrX 370473 106 - 22422827 ACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGGAAUUAGAACCGCGAGCAAGCGGAGCUCACAGUGACCGUAAC- ...((((((((....)))))))).-...((((.......))))(((((((.(((....))).))))....((..((((......))))..)).........)))...- ( -27.90, z-score = -1.94, R) >droSim1.chrX 269312 106 - 17042790 ACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGGAAUUAGAACCGCGAGCAAGCGGAGCUCACAGUGACCCCUAC- ...((((((((....)))))))).-...((((.......))))((.((((.(((....))).))))....((..((((......))))..))....)).........- ( -26.70, z-score = -1.92, R) >droSec1.super_10 228198 106 - 3036183 ACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGGAAUUAGAACCGCGAGCAAGCGGAGCUCACAGUGACCCCUAC- ...((((((((....)))))))).-...((((.......))))((.((((.(((....))).))))....((..((((......))))..))....)).........- ( -26.70, z-score = -1.92, R) >droYak2.chrX 299692 106 - 21770863 ACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGAGUGCUGAUGCGAUCGGAAUUAGAACCGCGAGCAAGCGGAGCUCAUAGUGACCCCCAC- ...((((((((....)))))))).-...((((.......))))...((((.(((....))).)))).((.((..((((......))))..)).)).(((.....)))- ( -27.10, z-score = -1.88, R) >droEre2.scaffold_4644 317917 106 - 2521924 ACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGAGUGCUGAUGCGAUCGGAAUCAGAACCGCGUGCAAGCGGAGCUUAUAGUGACCCCCAC- ...((((((((....)))))))).-.......((........((..((((.(((....))).))))..))....((((......)))).)).....(((.....)))- ( -27.40, z-score = -1.60, R) >droVir3.scaffold_13042 1512614 96 + 5191987 -----------AAAAAUUCAAAUAUAAAUCAUGAAAUUCUUAGCAAACAACUGCUAAUGCACUCAAAUCACUUAUCAC-CGCUAGCACCGAGCACAAAACCGCACACC -----------....................(((.....((((((......)))))).....))).............-.(((.......)))............... ( -8.30, z-score = -0.65, R) >consensus ACCUUUUGUGUAAAGACACAAAGA_AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGGAAUUAGAACCGCGAGCAAGCGGAGCUCACAGUGACCCCCAC_ ...((((((((....)))))))).....((((.......))))...((((.(((....))).))))....((..((((......))))..))................ (-17.38 = -19.22 + 1.84)

| Location | 370,482 – 370,587 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.35253 |
| G+C content | 0.44974 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -17.47 |
| Energy contribution | -20.20 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811173 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 370482 105 + 22422827 ACUGUGAGCUCCGCUUGCUCGCGGUUCUAAUU---CCGAUCGCAUCAGCAGUCGGCGUCCAUUAAUUGCAUGGUUU-UCUUUGUGUCUUUACACAAAAGGUGGCCAAAA- (((((((((.......))))))))).......---(((((.((....)).)))))...............((((..-(((((((((....)))).)))))..))))...- ( -33.80, z-score = -2.80, R) >droSim1.chrX 269321 105 + 17042790 ACUGUGAGCUCCGCUUGCUCGCGGUUCUAAUU---CCGAUCGCAUCAGCAGUCGGCGUCCAUUAAUUGCAUGGUUU-UCUUUGUGUCUUUACACAAAAGGUGGCCAAAA- (((((((((.......))))))))).......---(((((.((....)).)))))...............((((..-(((((((((....)))).)))))..))))...- ( -33.80, z-score = -2.80, R) >droSec1.super_10 228207 105 + 3036183 ACUGUGAGCUCCGCUUGCUCGCGGUUCUAAUU---CCGAUCGCAUCAGCAGUCGGCGUCCAUUAAUUGCAUGGUUU-UCUUUGUGUCUUUACACAAAAGGUGGCCAAAA- (((((((((.......))))))))).......---(((((.((....)).)))))...............((((..-(((((((((....)))).)))))..))))...- ( -33.80, z-score = -2.80, R) >droYak2.chrX 299701 105 + 21770863 ACUAUGAGCUCCGCUUGCUCGCGGUUCUAAUU---CCGAUCGCAUCAGCACUCGGCGUCCAUUAAUUGCAUGGUUU-UCUUUGUGUCUUUACACAAAAGGUGGCCAAAA- ...(((..(.(((..((((.((((((......---..))))))...))))..))).)..)))........((((..-(((((((((....)))).)))))..))))...- ( -30.20, z-score = -2.21, R) >droEre2.scaffold_4644 317926 106 + 2521924 ACUAUAAGCUCCGCUUGCACGCGGUUCUGAUU---CCGAUCGCAUCAGCACUCGGCGUCCAUUAAUUGCAUGGUUU-UCUUUGUGUCUUUACACAAAAGGUGGCCAAAAA .......((.((((......))))....(((.---((((..((....))..)))).)))........)).((((..-(((((((((....)))).)))))..)))).... ( -28.90, z-score = -1.65, R) >droVir3.scaffold_13042 1512622 88 - 5191987 GUUUUGUGCUCGG--UGCUAGCGGUGAUAAGUGAUUUGAGUGCAUUAGCAGUUGUUUGCUAAGAAUUUCAUGAUUUAUAUUUGAAUUUUU-------------------- ....((..(((((--.....((........))...)))))..))(((((((....))))))).........((((((....))))))...-------------------- ( -17.30, z-score = 0.05, R) >consensus ACUGUGAGCUCCGCUUGCUCGCGGUUCUAAUU___CCGAUCGCAUCAGCAGUCGGCGUCCAUUAAUUGCAUGGUUU_UCUUUGUGUCUUUACACAAAAGGUGGCCAAAA_ .....((((.......))))((((((.........((((..((....))..))))........)))))).(((((....((((((......))))))....))))).... (-17.47 = -20.20 + 2.72)

| Location | 370,482 – 370,587 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.35253 |
| G+C content | 0.44974 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -16.23 |
| Energy contribution | -18.45 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671842 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
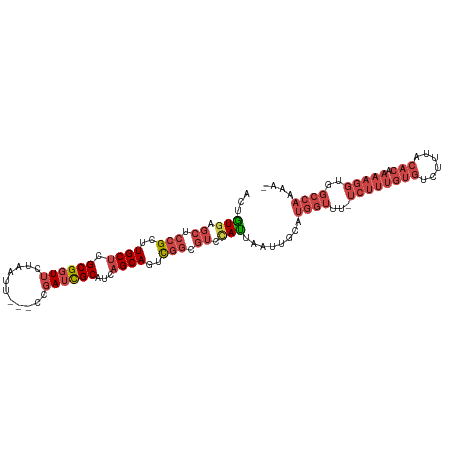
>dm3.chrX 370482 105 - 22422827 -UUUUGGCCACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGG---AAUUAGAACCGCGAGCAAGCGGAGCUCACAGU -..(((((....((((((((....)))))))).-...((((.......))))..)))))(((.(((.((..((..---.....)).((((......)))).)))))))). ( -30.10, z-score = -2.12, R) >droSim1.chrX 269321 105 - 17042790 -UUUUGGCCACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGG---AAUUAGAACCGCGAGCAAGCGGAGCUCACAGU -..(((((....((((((((....)))))))).-...((((.......))))..)))))(((.(((.((..((..---.....)).((((......)))).)))))))). ( -30.10, z-score = -2.12, R) >droSec1.super_10 228207 105 - 3036183 -UUUUGGCCACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGG---AAUUAGAACCGCGAGCAAGCGGAGCUCACAGU -..(((((....((((((((....)))))))).-...((((.......))))..)))))(((.(((.((..((..---.....)).((((......)))).)))))))). ( -30.10, z-score = -2.12, R) >droYak2.chrX 299701 105 - 21770863 -UUUUGGCCACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGAGUGCUGAUGCGAUCGG---AAUUAGAACCGCGAGCAAGCGGAGCUCAUAGU -....(((....((((((((....)))))))).-...((((.......))))..)))((((.(((.(((...(((---........)))...)))..))).))))..... ( -29.70, z-score = -1.87, R) >droEre2.scaffold_4644 317926 106 - 2521924 UUUUUGGCCACCUUUUGUGUAAAGACACAAAGA-AAACCAUGCAAUUAAUGGACGCCGAGUGCUGAUGCGAUCGG---AAUCAGAACCGCGUGCAAGCGGAGCUUAUAGU ....(((.....((((((((....)))))))).-...))).((........((..((((.(((....))).))))---..))....((((......)))).))....... ( -28.70, z-score = -1.14, R) >droVir3.scaffold_13042 1512622 88 + 5191987 --------------------AAAAAUUCAAAUAUAAAUCAUGAAAUUCUUAGCAAACAACUGCUAAUGCACUCAAAUCACUUAUCACCGCUAGCA--CCGAGCACAAAAC --------------------....................(((.....((((((......)))))).....)))..............(((....--...)))....... ( -8.30, z-score = -1.03, R) >consensus _UUUUGGCCACCUUUUGUGUAAAGACACAAAGA_AAACCAUGCAAUUAAUGGACGCCGACUGCUGAUGCGAUCGG___AAUUAGAACCGCGAGCAAGCGGAGCUCACAGU ............((((((((....)))))))).....((((.......))))...((((.(((....))).))))...........((((......)))).......... (-16.23 = -18.45 + 2.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:45 2011