| Sequence ID | dm3.chrX |
|---|---|
| Location | 352,240 – 352,392 |
| Length | 152 |
| Max. P | 0.996633 |

| Location | 352,240 – 352,352 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Shannon entropy | 0.20998 |
| G+C content | 0.38504 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.49 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 352240 112 + 22422827 UGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGUUUUUAAAUAGAAACUCGCUAUUUCCCCAUGU-AUUGUUUAAAAGAGGUAU- ...........((((.((------.....((((((((((.(((((((((..(((((((.(.......).)))))))..))))))))).)))....)))))-..))......)).)))).- ( -21.40, z-score = -1.38, R) >droSim1.chrX 259267 109 + 17042790 UGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUC----AUUUCAAAGCGGCAU- ((((((((((((((..((------......)).)))))))))(((((((..(((((((((.......)))))))))..)))))))...............----.))))).........- ( -28.20, z-score = -3.25, R) >droSec1.super_10 209908 109 + 3036183 UGAAAACAAUUUACCACU------CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUC----AUUUCAAAGCGGCAU- ((((((((((((((..((------......)).)))))))))(((((((..(((((((((.......)))))))))..)))))))...............----.))))).........- ( -28.20, z-score = -3.25, R) >droYak2.chrX 279494 114 + 21770863 UGAAAACAAUUUACCACU------CCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAGAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUCAAAGCGAACGA (((((.((((......((------((.....).)))(((.(((((((((.((((((((((.......)))))))))).))))))))).)))..........))))))))).......... ( -32.00, z-score = -3.46, R) >droEre2.scaffold_4644 300009 119 + 2521924 UGAAAACAAUUUACCACUGCGAUCCCGAUCAGCGAGAAUUGUGAGUUUCACUUUAAAGUCGAACGCAGACUUUGAAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUUAAAACGGCAU- ..((((((((........(((((....))).))(..(((.(((((((((.((((((((((.......)))))))))).))))))))).)))..).......))))))))..........- ( -35.20, z-score = -3.96, R) >consensus UGAAAACAAUUUACCACU______CCGAUCAGCGUGAAUUGUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAUUU_AUUGUUUCAAAGCGGCAU_ (((((((((((((((................).)))))))))(((((((..(((((((((.......)))))))))..)))))))....................))))).......... (-23.29 = -23.49 + 0.20)



| Location | 352,240 – 352,352 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Shannon entropy | 0.20998 |
| G+C content | 0.38504 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.83 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 352240 112 - 22422827 -AUACCUCUUUUAAACAAU-ACAUGGGGAAAUAGCGAGUUUCUAUUUAAAAACUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCA -...........(((((((-......((((((.....))))))........((..(.(((((..(((..(((((........)))))..)))))))------))..))..)))))))... ( -19.90, z-score = -0.52, R) >droSim1.chrX 259267 109 - 17042790 -AUGCCGCUUUGAAAU----GAAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCA -.(((((((((((...----....(.(..(((.(.(((((((..(((((((((.......)))))))))..))))))).).)))..).)...))))------)))))))........... ( -34.00, z-score = -3.39, R) >droSec1.super_10 209908 109 - 3036183 -AUGCCGCUUUGAAAU----GAAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG------AGUGGUAAAUUGUUUUCA -.(((((((((((...----....(.(..(((.(.(((((((..(((((((((.......)))))))))..))))))).).)))..).)...))))------)))))))........... ( -34.00, z-score = -3.39, R) >droYak2.chrX 279494 114 - 21770863 UCGUUCGCUUUGAAACAAUGAAAUGGGGAAAUGGCGAGUUUCCCUCUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCUCGCUGAUCGG------AGUGGUAAAUUGUUUUCA ....(((((((((..((......))(((((.((..(((((((.((.(((((((.......))))))).)).))))))).)).))))).....))))------)))))............. ( -32.70, z-score = -2.38, R) >droEre2.scaffold_4644 300009 119 - 2521924 -AUGCCGUUUUAAAACAAUGAAAUGGGGAAAUGGCGAGUUUCCCUUCAAAGUCUGCGUUCGACUUUAAAGUGAAACUCACAAUUCUCGCUGAUCGGGAUCGCAGUGGUAAAUUGUUUUCA -..((((((((....((......))..))))))))(((((((.(((.((((((.......)))))).))).)))))))((((((.((((((..((....))))))))..))))))..... ( -35.40, z-score = -2.47, R) >consensus _AUGCCGCUUUGAAACAAU_AAAUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCACAAUUCACGCUGAUCGG______AGUGGUAAAUUGUUUUCA ..(((((((...............(.(..(((.(.(((((((..(((((((((.......)))))))))..))))))).).)))..).).............)))))))........... (-22.87 = -23.83 + 0.96)

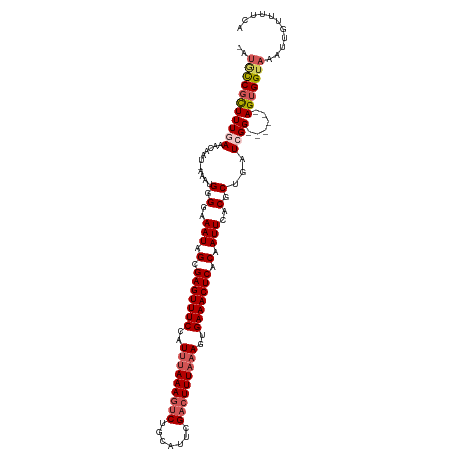
| Location | 352,274 – 352,392 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.43232 |
| G+C content | 0.41268 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.996633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
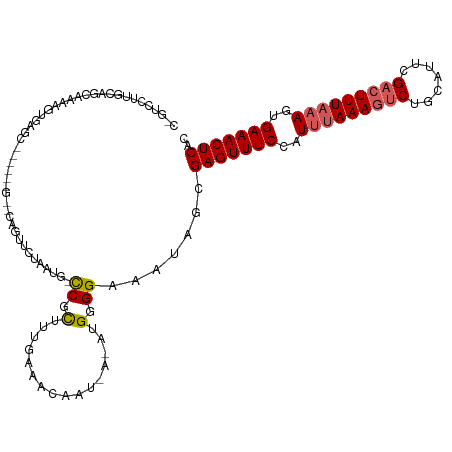
>dm3.chrX 352274 118 + 22422827 GUGAGUUUCACUUUAAAGUCGAAUGCAGUUUUUAAAUAGAAACUCGCUAUUUCCCCAUGU-AUUGUUUAAAAGAGG-UAUUAAAACAGCCCGCACCCGCUCAUUUUUGCUGCAAGGACGG (((((((((..(((((((.(.......).)))))))..))))))))).......((.(((-(..((..(((.((((-(.........))).((....)))).)))..)))))).)).... ( -22.30, z-score = -0.03, R) >droSim1.chrX 259301 106 + 17042790 GUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAU----UCAUUUCAAAGCGG-CAUUACAACUG---------GCUCGCUUUUGCUGCAAGGACAG (((((((((..(((((((((.......)))))))))..)))))))))...........----..........((((-((.........---------.........))))))........ ( -30.47, z-score = -2.94, R) >droSec1.super_10 209942 106 + 3036183 GUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAU----UCAUUUCAAAGCGG-CAUUACAACUG---------GCUCGCUUUUGCUGCAAGGACAG (((((((((..(((((((((.......)))))))))..)))))))))...........----..........((((-((.........---------.........))))))........ ( -30.47, z-score = -2.94, R) >droYak2.chrX 279528 101 + 21770863 GUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAGAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUCAAAGCGAACGACAGGAUUAGUC-------AUUACUUUUA------------ (((((((((.((((((((((.......)))))))))).)))))))))...............(((((....)))))..(((.......)))-------..........------------ ( -28.60, z-score = -3.71, R) >droEre2.scaffold_4644 300049 107 + 2521924 GUGAGUUUCACUUUAAAGUCGAACGCAGACUUUGAAGGGAAACUCGCCAUUUCCCCAUUUCAUUGUUUUAAAACGG-CAUUAGAACUGGUCGCAGCCACUCACUUUUG------------ (((((((((.((((((((((.......)))))))))).))))))))).................(((((((.....-..)))))))((((....))))..........------------ ( -30.70, z-score = -3.15, R) >consensus GUGAGUUUCACUUUAAAGUCGAAUGCAGACUUUAAAUGGAAACUCGCUAUUUCCCCAU_U_AUUGUUUCAAAGCGG_CAUUACAACUG__C______GCUCACUUUUGCUGCAAGGAC_G (((((((((..(((((((((.......)))))))))..)))))))))......................................................................... (-20.90 = -20.62 + -0.28)

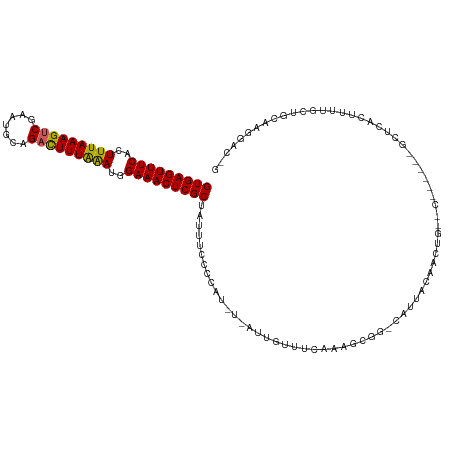
| Location | 352,274 – 352,392 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.43232 |
| G+C content | 0.41268 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.79 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 352274 118 - 22422827 CCGUCCUUGCAGCAAAAAUGAGCGGGUGCGGGCUGUUUUAAUA-CCUCUUUUAAACAAU-ACAUGGGGAAAUAGCGAGUUUCUAUUUAAAAACUGCAUUCGACUUUAAAGUGAAACUCAC (((..(((((...........)))))..)))((((((((....-((.............-....)).))))))))(((((((..((((((.............))))))..))))))).. ( -30.05, z-score = -1.88, R) >droSim1.chrX 259301 106 - 17042790 CUGUCCUUGCAGCAAAAGCGAGC---------CAGUUGUAAUG-CCGCUUUGAAAUGA----AUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCAC ...(((((.((...((((((.((---------..........)-)))))))....)).----..)))))......(((((((..(((((((((.......)))))))))..))))))).. ( -31.80, z-score = -2.64, R) >droSec1.super_10 209942 106 - 3036183 CUGUCCUUGCAGCAAAAGCGAGC---------CAGUUGUAAUG-CCGCUUUGAAAUGA----AUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCAC ...(((((.((...((((((.((---------..........)-)))))))....)).----..)))))......(((((((..(((((((((.......)))))))))..))))))).. ( -31.80, z-score = -2.64, R) >droYak2.chrX 279528 101 - 21770863 ------------UAAAAGUAAU-------GACUAAUCCUGUCGUUCGCUUUGAAACAAUGAAAUGGGGAAAUGGCGAGUUUCCCUCUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCAC ------------..((((((((-------(((.......)))))).)))))........................(((((((.((.(((((((.......))))))).)).))))))).. ( -26.60, z-score = -3.02, R) >droEre2.scaffold_4644 300049 107 - 2521924 ------------CAAAAGUGAGUGGCUGCGACCAGUUCUAAUG-CCGUUUUAAAACAAUGAAAUGGGGAAAUGGCGAGUUUCCCUUCAAAGUCUGCGUUCGACUUUAAAGUGAAACUCAC ------------.....((((((.(((.((.(((.((((....-((((((((......)))))))))))).))))))))(((.(((.((((((.......)))))).))).))))))))) ( -31.00, z-score = -2.39, R) >consensus C_GUCCUUGCAGCAAAAGUGAGC______G__CAGUUCUAAUG_CCGCUUUGAAACAAU_A_AUGGGGAAAUAGCGAGUUUCCAUUUAAAGUCUGCAUUCGACUUUAAAGUGAAACUCAC ............................................((.(................).)).......(((((((..(((((((((.......)))))))))..))))))).. (-15.31 = -15.79 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:43 2011