
| Sequence ID | dm3.chrX |
|---|---|
| Location | 342,673 – 342,734 |
| Length | 61 |
| Max. P | 0.765781 |

| Location | 342,673 – 342,734 |
|---|---|
| Length | 61 |
| Sequences | 12 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 90.57 |
| Shannon entropy | 0.20816 |
| G+C content | 0.38413 |
| Mean single sequence MFE | -8.41 |
| Consensus MFE | -7.60 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 342673 61 + 22422827 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUAGAGACGAG------- (((....)))....((....((((......))))........(((......)))....)).------- ( -7.90, z-score = -1.38, R) >droSim1.chrX 249799 61 + 17042790 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUAGAGACGAG------- (((....)))....((....((((......))))........(((......)))....)).------- ( -7.90, z-score = -1.38, R) >droSec1.super_10 200400 61 + 3036183 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUAGAGACGAG------- (((....)))....((....((((......))))........(((......)))....)).------- ( -7.90, z-score = -1.38, R) >droYak2.chrX 269849 67 + 21770863 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUAGAGACGAAGACGAG- .((((..(((..........((((......))))........(((......))).).))..))))..- ( -9.10, z-score = -1.54, R) >droEre2.scaffold_4644 290505 61 + 2521924 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUAGAGACGAG------- (((....)))....((....((((......))))........(((......)))....)).------- ( -7.90, z-score = -1.38, R) >droAna3.scaffold_13117 5067817 61 + 5790199 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAGACGAG------- (((....)))....((....((((......))))..................(....))).------- ( -9.50, z-score = -1.64, R) >dp4.chrXL_group1e 4347236 54 + 12523060 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGG-------------- (((....)))..........((((......))))....................-------------- ( -7.60, z-score = -1.53, R) >droPer1.super_17 1289746 54 + 1930428 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGG-------------- (((....)))..........((((......))))....................-------------- ( -7.60, z-score = -1.53, R) >droWil1.scaffold_181096 7211374 68 - 12416693 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAAACGGAAGACGAA (((....)))....((....((((......)))).................((....))......)). ( -9.80, z-score = -1.42, R) >droVir3.scaffold_13042 1467655 61 - 5191987 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAGACUCG------- (((....)))..........((((......))))..................(....)...------- ( -8.60, z-score = -1.03, R) >droMoj3.scaffold_6308 2417407 61 - 3356042 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAGACGAG------- (((....)))....((....((((......))))..................(....))).------- ( -9.50, z-score = -1.64, R) >droGri2.scaffold_14853 7530190 61 - 10151454 GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAGGCUCC------- (((....)))..........((((......))))...........................------- ( -7.60, z-score = -0.09, R) >consensus GCGUUGACGCAAAAUCAAAAGAUCACACAGGAUCAAUUACAACUAAUGAUAUGGAGACGAG_______ (((....)))..........((((......)))).................................. ( -7.60 = -7.60 + 0.00)

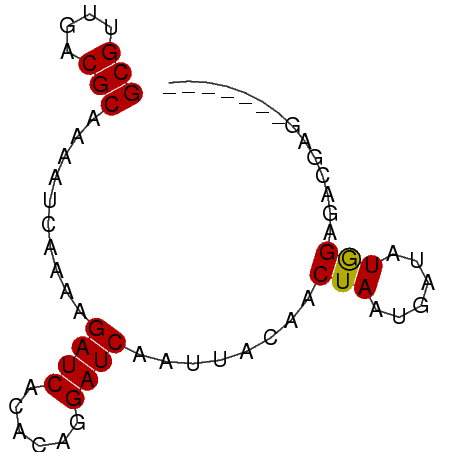

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:40 2011