| Sequence ID | dm3.chrX |
|---|---|
| Location | 316,188 – 316,289 |
| Length | 101 |
| Max. P | 0.527156 |

| Location | 316,188 – 316,289 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Shannon entropy | 0.45289 |
| G+C content | 0.43204 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 316188 101 - 22422827 AACAAAUCAAACUGCCAGUACAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUCGAGUGGU------------------UGGCGCAAGUGAAAUAAUUUGCUUG .............(((.(((((((...(....).(....)..))))))).....(((((((........).)))))------------------))))((((((......))))))... ( -28.60, z-score = -2.67, R) >droPer1.super_17 1265021 112 - 1930428 AACAAAUCAAACUGCCAUUGCAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGGGACCACCAGAGAGCUCUCUGGU-------UCUCUGGUUUGUGGCGCAAGUGAAAUAAUUUGCUGG .............(((((((((((...(....).(....)..))))))..........(((((((((((....)))-------))))))))..)))))((((((......))))))... ( -35.40, z-score = -2.28, R) >dp4.chrXL_group1e 4322608 112 - 12523060 AACAAAUCAAACUGCCAUUGCAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGGGACCACCAGAGAGAUCUCUGGU-------UCUCUGGUUUGUGGCGCAAGUGAAAUAAUUUGCUGG .((((((((.....(((.((((((...(....).(....)..))))))...)))((..(((((((....)))))))-------..))))))))))...((((((......))))))... ( -34.70, z-score = -2.37, R) >droSim1.chrX 228223 101 - 17042790 AACAAAUCAAACUGCCAGUACAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUCAAGUGGU------------------UGGCGCAAGUGAAAUAAUUUGCUGG .............(((.(((((((...(....).(....)..))))))).....((((((...........)))))------------------))))((((((......))))))... ( -27.60, z-score = -2.26, R) >droSec1.super_10 178343 101 - 3036183 AACAAAUCAAACUGCCAGUACAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUCGAGUGGU------------------UGGCGCAAGUGAAAUAAUUUGCUGG .............(((.(((((((...(....).(....)..))))))).....(((((((........).)))))------------------))))((((((......))))))... ( -28.60, z-score = -2.50, R) >droYak2.chrX 248692 101 - 21770863 AACAAAUCAAACUGCCAGUGCAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUCGAGUGGU------------------UGGCGCAAGUGAAAUAAUUUGCUGG .............(((.(((((((...(....).(....)..))))))).....(((((((........).)))))------------------))))((((((......))))))... ( -28.60, z-score = -1.96, R) >droEre2.scaffold_4644 267974 101 - 2521924 AACAAAUCAAACUGCCAGUACAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUCGAGUGGU------------------UGGCGCAAGUGAAAUAAUUUGCUGG .............(((.(((((((...(....).(....)..))))))).....(((((((........).)))))------------------))))((((((......))))))... ( -28.60, z-score = -2.50, R) >droWil1.scaffold_181096 7186720 91 + 12416693 AACAAAUCAAACUGCCAUUGAAGUAGAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGAGUGAU----------------------------CGUAAGUGAAAUAAUUUGCUGG ......((...((((.......))))..))..(((....((((((((.((((..((.(((....))).)----------------------------)...))))..))))))))))). ( -14.30, z-score = 0.02, R) >droAna3.scaffold_13117 5042250 103 - 5790199 AACAAAUCAAACUGCCAGUACGGGAGAGGAAACAGAAAUCAAGUUGUACUUUCAGACCACCA----------------GCAGAUCGAUACAACUGGGCGCAAGUGAAAUAAUUUGCUGG ...........(((..(((((((....(....).(....)...)))))))..)))....(((----------------((((((.......(((.......)))......))))))))) ( -22.32, z-score = -0.79, R) >droVir3.scaffold_13042 1411498 118 + 5191987 AACAAAUCAAACUGCCAGCGCAGUAAAGGAAACAGAAAUCAAGUUGUACUUGAAGACCACCGGAGAGUCCAGCAGUCCGCCAGGCCGGGCGAUU-GGCGCAAGUGAAAUAAUUUGCUGG ......((..(((((....)))))...(....).))..(((((.....)))))...............(((((((..((((......)))).((-.((....)).)).....))))))) ( -31.90, z-score = -0.61, R) >droMoj3.scaffold_6308 2374072 93 + 3356042 AACAAAUCAAACUGCCAGUGCAGCAGAGGAAACAGAAAUCAAGUUGUACUUGGAGACCA-------------------------CCAGGCGAUU-GGCGCAAGUGAAAUAAUUUGCUGG .............(((((((((((...(....).(....)..))))).(((((......-------------------------)))))..)))-)))((((((......))))))... ( -24.60, z-score = -1.37, R) >droGri2.scaffold_14853 7497863 96 + 10151454 AACAAAUCAAACUGCCAUUGCAGCAAAGGAAACAGAAAUCAAGUUGUACUUGGAGACCACC----------------------GCGUAGCGAUU-GGCGCAAGUGAAAUAAUUUGCUGG .............((((((((.((...(....).....(((((.....)))))........----------------------))...)))).)-)))((((((......))))))... ( -24.20, z-score = -1.17, R) >consensus AACAAAUCAAACUGCCAGUGCAGCAAAGGAAACAGAAAUCAAGUUGUACAUUGAGACCACCAGCAGUUC_AGUGGU__________________UGGCGCAAGUGAAAUAAUUUGCUGG .............(((..((((((...(....).(....)..))))))(.....)........................................)))((((((......))))))... (-12.58 = -12.48 + -0.10)

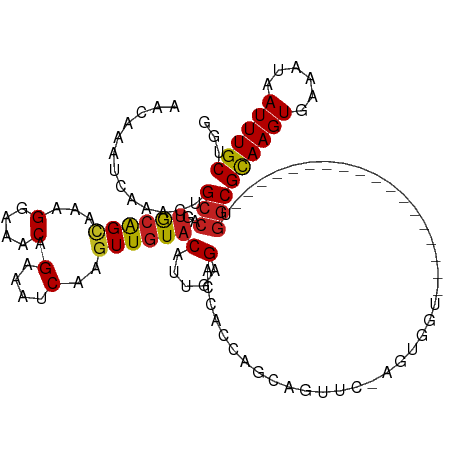

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:37 2011