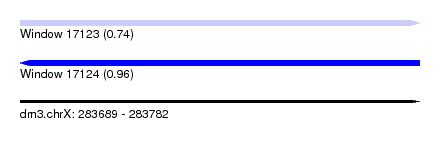
| Sequence ID | dm3.chrX |
|---|---|
| Location | 283,689 – 283,782 |
| Length | 93 |
| Max. P | 0.960498 |

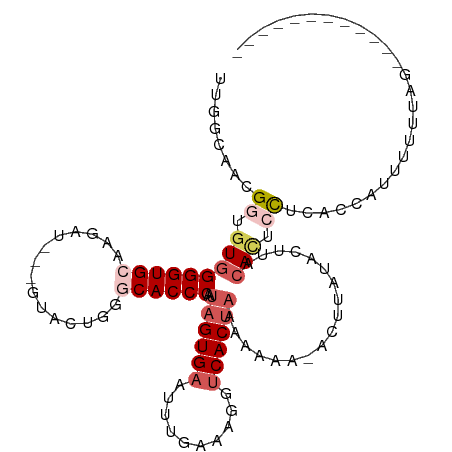
| Location | 283,689 – 283,782 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.62 |
| Shannon entropy | 0.52328 |
| G+C content | 0.41487 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -12.32 |
| Energy contribution | -13.96 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 283689 93 + 22422827 ------UUGGGAUGGGGUGCAAGAU---GUACUGGGCACCCAAUAGUGAAUUUGUAGGGUCACAAAAAAAA-ACUUAUUCUUACACUCCUCACCAUUUUUUUG------------ ------.(((((.((((((.((((.---....(((....)))...((((..........))))........-......)))).)))))))).)))........------------ ( -22.60, z-score = -0.67, R) >droEre2.scaffold_4644 234672 114 + 2521924 UUGGCGACGGUGUGGGGUGGACGACGACGUACUGGGCACCCAAUAGUGAAUUUGAAAGGUCACUAAAAAGAUACUUA-ACUACCAUUGUUGGUAGUAUUUGAUACAAUUUUUGGG ...((..(((((((..((.....))..))))))).)).(((((((((((...(....).))))))............-(((((((....)))))))..............))))) ( -29.20, z-score = -0.90, R) >droYak2.chrX 216340 112 + 21770863 UUGGCAACGGUGUGGGGUGGAAGAU---GCACUGGGCACCCAAUAGUGAAUUUGAAUAGCCACUCAAAAGAUACUUAUACUGCCACUAUUGGUAAUAUAUAAUACACUUUUUGGA ..(....)((((((((((((.((((---.((((((....))...)))).))))......))))))..........((((.(((((....))))).))))...))))))....... ( -27.20, z-score = -0.57, R) >droSec1.super_10 145502 90 + 3036183 UUGAGAACGGUGUGGGGUGCAAGAU---GUACUGGGCACCCAAUAGUGAAUUUCAAAGGUCACUAAAUAA---CUUA-------ACUCCACACCAUUUUUUGG------------ ..(((((.((((((((((..(((.(---((..(((....))).((((((..........)))))).))).---))).-------)))))))))).)))))...------------ ( -29.50, z-score = -2.93, R) >droSim1.chrX 195963 97 + 17042790 UUGGGAACGGUGUGGGGUGCAAGAU---GUACUGGGCACCCAAUAGUGAAUUUCAAAGGUCACUAAAUAA---CUUAUUCUUACACUCCACACCAUUUUUUGG------------ (..((((.(((((((((((.((((.---....(((....))).((((((..........)))))).....---.....)))).)))))))))))..))))..)------------ ( -32.80, z-score = -3.36, R) >consensus UUGGCAACGGUGUGGGGUGCAAGAU___GUACUGGGCACCCAAUAGUGAAUUUGAAAGGUCACUAAAAAAA_ACUUAUACUUACACUCCUCACCAUUUUUUAG____________ ........((.(((((((((.((........))..))))))..((((((..........))))))..................))).)).......................... (-12.32 = -13.96 + 1.64)

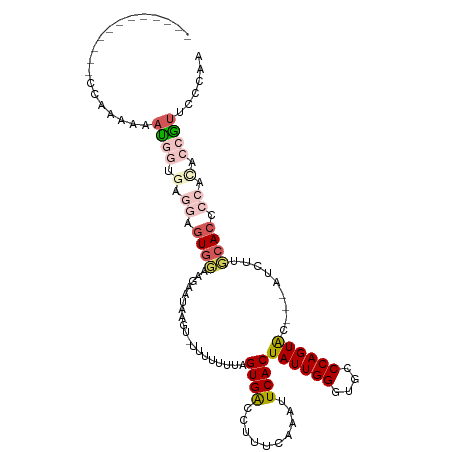
| Location | 283,689 – 283,782 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.62 |
| Shannon entropy | 0.52328 |
| G+C content | 0.41487 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -10.46 |
| Energy contribution | -12.22 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 283689 93 - 22422827 ------------CAAAAAAAUGGUGAGGAGUGUAAGAAUAAGU-UUUUUUUUGUGACCCUACAAAUUCACUAUUGGGUGCCCAGUAC---AUCUUGCACCCCAUCCCAA------ ------------........(((.((((.((((((((......-.....((((((....)))))).....((((((....)))))).---.)))))))).)).))))).------ ( -26.80, z-score = -2.89, R) >droEre2.scaffold_4644 234672 114 - 2521924 CCCAAAAAUUGUAUCAAAUACUACCAACAAUGGUAGU-UAAGUAUCUUUUUAGUGACCUUUCAAAUUCACUAUUGGGUGCCCAGUACGUCGUCGUCCACCCCACACCGUCGCCAA ..........((((.....(((((((....)))))))-...((((((...((((((..........))))))..))))))...))))(.((.((............)).)).).. ( -22.00, z-score = -1.66, R) >droYak2.chrX 216340 112 - 21770863 UCCAAAAAGUGUAUUAUAUAUUACCAAUAGUGGCAGUAUAAGUAUCUUUUGAGUGGCUAUUCAAAUUCACUAUUGGGUGCCCAGUGC---AUCUUCCACCCCACACCGUUGCCAA ..(((...((((...........(((((((((((.......))....(((((((....)))))))..)))))))))((((.....))---))..........))))..))).... ( -21.90, z-score = -0.16, R) >droSec1.super_10 145502 90 - 3036183 ------------CCAAAAAAUGGUGUGGAGU-------UAAG---UUAUUUAGUGACCUUUGAAAUUCACUAUUGGGUGCCCAGUAC---AUCUUGCACCCCACACCGUUCUCAA ------------......(((((((((....-------....---.....((((((..........))))))..((((((..((...---..)).)))))))))))))))..... ( -26.70, z-score = -2.75, R) >droSim1.chrX 195963 97 - 17042790 ------------CCAAAAAAUGGUGUGGAGUGUAAGAAUAAG---UUAUUUAGUGACCUUUGAAAUUCACUAUUGGGUGCCCAGUAC---AUCUUGCACCCCACACCGUUCCCAA ------------......((((((((((.((((((((.....---......(((((..........)))))(((((....)))))..---.)))))))).))))))))))..... ( -35.70, z-score = -5.21, R) >consensus ____________CCAAAAAAUGGUGAGGAGUGGAAGAAUAAGU_UUUUUUUAGUGACCUUUCAAAUUCACUAUUGGGUGCCCAGUAC___AUCUUGCACCCCACACCGUUCCCAA ...................((((.(.((.((((...................((((..........))))((((((....)))))).........)))).)).).))))...... (-10.46 = -12.22 + 1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:35 2011