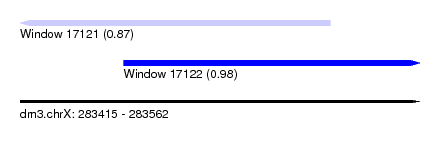
| Sequence ID | dm3.chrX |
|---|---|
| Location | 283,415 – 283,562 |
| Length | 147 |
| Max. P | 0.980107 |

| Location | 283,415 – 283,529 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Shannon entropy | 0.31213 |
| G+C content | 0.40497 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -17.51 |
| Energy contribution | -20.01 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
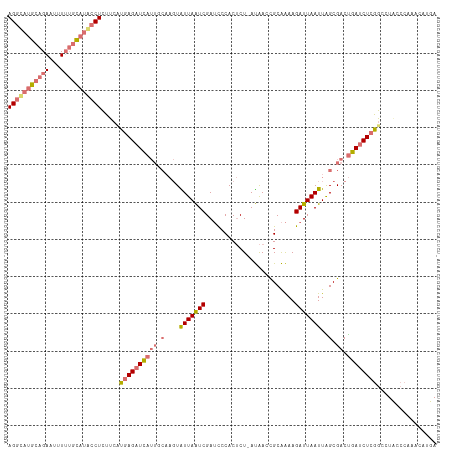
>dm3.chrX 283415 114 - 22422827 AGACAUGCAGAAUUUGUGCAUACCUCUUCAUGAGAUCAUUUCAAGUGUUAAUCGAUCCCACUCU-AUAACCGCAAAAGAUUAAUUGGCGACUGAUCUCAGCCUCCCCUAACAUGA ....(((((.......))))).........((((((((.......(((((((..(((.......-............)))..)))))))..))))))))................ ( -18.71, z-score = 0.11, R) >droYak2.chrX 216057 108 - 21770863 AGGCAUGCAGAAUUUUUCUUAGCCUAUGCAUCAGUUUA-------UGUUAGUCGAGCCCACACUAACGACUGCGGAAGAUUAAUUGGCGACCGAUCUCGUUUUCCCCAAACAUGA ((((.((.(((.....)))))))))..........(((-------(((((((((((......))..)))))(.(((((((..(((((...)))))...))))))).).))))))) ( -22.00, z-score = 0.03, R) >droSec1.super_10 145206 114 - 3036183 AGGUAUGCAGAAUUUUUGCAUACCUCUUCUUGAGAUCAUUGCAAGUAUUAAUCGAUUCCACUCU-AUAACCGCAAAAGAUUAAUUAGCGACUGAUCUCGGCCUACCCAAACAUUA (((((((((((...)))))))))))......(((((((((((....(((((((...........-............)))))))..)))).)))))))((.....))........ ( -30.40, z-score = -4.35, R) >droSim1.chrX 195689 114 - 17042790 AGGUAUGCAGAAUUUUAGCAUACCUCUUCUUGAGAUCAUUGCAAGUAUUAAUCGAUCCCACUCU-AUAACCGCAAAAGAUUAAUUAGCGACUGAUCUCGGCCUACCCAAACAUUA ((((((((.........))))))))......(((((((((((....(((((((...........-............)))))))..)))).)))))))((.....))........ ( -27.70, z-score = -3.48, R) >consensus AGGCAUGCAGAAUUUUUGCAUACCUCUUCAUGAGAUCAUUGCAAGUAUUAAUCGAUCCCACUCU_AUAACCGCAAAAGAUUAAUUAGCGACUGAUCUCGGCCUACCCAAACAUGA (((((((((((...))))))))))).....((((((((((((....(((((((........................)))))))..)))).))))))))................ (-17.51 = -20.01 + 2.50)

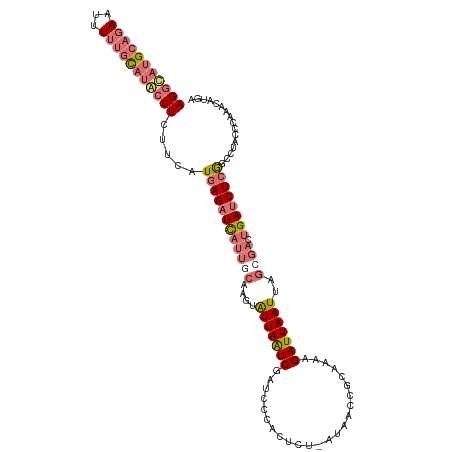
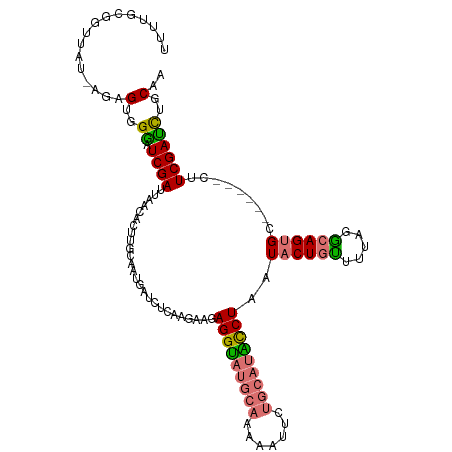
| Location | 283,453 – 283,562 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Shannon entropy | 0.35514 |
| G+C content | 0.39450 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 283453 109 + 22422827 UUUUGCGGUUAU-AGAGUGGGAUCGAUUAACACUUGAAAUGAUCUCAUGAAGAGGUAUGCACAAAUUCUGCAUGUCUAAUCCUGUUUUAGACAGUGC------CUUCGAUCUGCAA ..((((((....-...(((((((((.((........)).)))))))))...((((((((((.......))).(((((((.......)))))))))))------)))....)))))) ( -30.50, z-score = -1.91, R) >droYak2.chrX 216095 109 + 21770863 UUCCGCAGUCGUUAGUGUGGGCUCGACUAACA-------UAAACUGAUGCAUAGGCUAAGAAAAAUUCUGCAUGCCUAAUACUGCAUUGGGAAGUGCUUUAGACUUCGACUUGCAA ....((((((((((((.((....)))))))).-------....(..((((((((((..(((.....)))....)))))....)))))..)(((((.......)))))))).))).. ( -33.50, z-score = -2.43, R) >droSec1.super_10 145244 109 + 3036183 UUUUGCGGUUAU-AGAGUGGAAUCGAUUAAUACUUGCAAUGAUCUCAAGAAGAGGUAUGCAAAAAUUCUGCAUACCUAAUACUGUUUUAGGCAGUGU------CUUCGAUCUGCGA ..((((((((((-.(((((...........)))))...))))))....(((((((((((((.......))))))))).(((((((.....)))))))------)))).....)))) ( -31.50, z-score = -2.64, R) >droSim1.chrX 195727 109 + 17042790 UUUUGCGGUUAU-AGAGUGGGAUCGAUUAAUACUUGCAAUGAUCUCAAGAAGAGGUAUGCUAAAAUUCUGCAUACCUAAUACUGUUUUAGGCAGUGU------CUUCGAUCUGCAA ..((((((....-....((((((((..............)))))))).((((((((((((.........)))))))).(((((((.....)))))))------))))...)))))) ( -33.94, z-score = -3.36, R) >consensus UUUUGCGGUUAU_AGAGUGGGAUCGAUUAACACUUGCAAUGAUCUCAAGAAGAGGUAUGCAAAAAUUCUGCAUACCUAAUACUGUUUUAGGCAGUGC______CUUCGAUCUGCAA ................(..((.((((..........................(((((((((.......)))))))))..((((((.....)))))).........))))))..).. (-20.24 = -20.43 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:34 2011