| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,411,742 – 9,411,841 |
| Length | 99 |
| Max. P | 0.616796 |

| Location | 9,411,742 – 9,411,841 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.60 |
| Shannon entropy | 0.42301 |
| G+C content | 0.50476 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -14.70 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
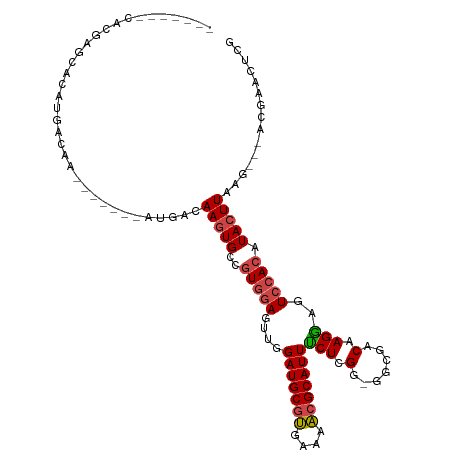
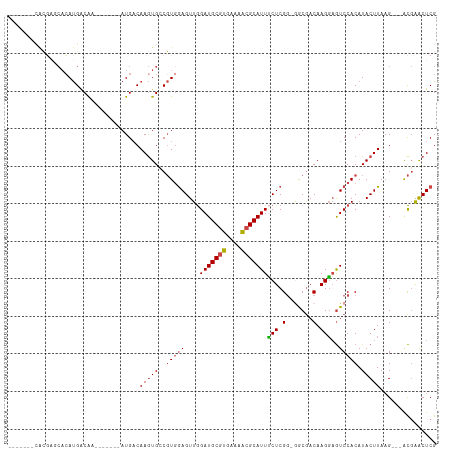
>dm3.chr2L 9411742 99 + 23011544 -------CACGGGCACAUGACAA----CAAAUGACAAGUGCCGUGGAGUUGGAUGCGUGAAAACGCAUUUCUCGC-GACGACAAGGAGUCCACAUACUUAAG---ACGAGCUCG -------(((((.(((.((.((.----....)).)).))))))))(((((.(((((((....)))))).....(.-(((........))))...........---.).))))). ( -30.70, z-score = -2.06, R) >droSim1.chr2L 9188093 96 + 22036055 -------CACGAGCACAUGACAA-------AUGACAAGUGCCGUGGAGUUGGAUGCGUGAAAACGCAUUUCUCGG-AGCGACAAGGAGUCCACAUACUUAAG---ACGAACUCG -------((((.((((.((....-------....)).))))))))(((((.(((((((....)))))).....((-(.(........))))...........---.).))))). ( -30.20, z-score = -2.56, R) >droSec1.super_3 4865398 96 + 7220098 -------CACGAGCACAUGACAA-------AUGACAAGUGCCGUGGAGUUGGAUGCGUGAAAACGCAUUUCUCGG-AGCGACAAGGAGUCCACAUACUUAAG---ACGAACUCG -------((((.((((.((....-------....)).))))))))(((((.(((((((....)))))).....((-(.(........))))...........---.).))))). ( -30.20, z-score = -2.56, R) >droYak2.chr2L 12083065 105 + 22324452 -----CACAUGACCACAUGACAAAAGACAAAUGACAAGUGCCGUCGAGUUGGAUGCGUGCAAACGCAUUUCUCGG-GACGACAAGGAGUCCACAUACUUAAG---GCGAACUCG -----..((((....)))).............((....((((..((((...(((((((....))))))).))))(-(((........))))..........)---)))...)). ( -29.00, z-score = -1.64, R) >droEre2.scaffold_4929 10020828 110 + 26641161 CACGAGCAGUUGACAAAUGACAAAUGACGAAUGCCAAGUGCCGUGGAGUUGGAUGCGUGAAAACGCAUUGCUCGG-GGCGACAAGCAGUCCACAUACUUAAG---ACGAGCUCG ((((.(((.(((.((..((........))..)).))).)))))))......(((((((....)))))))((((((-(((........)))).....(....)---.)))))... ( -34.60, z-score = -1.59, R) >droAna3.scaffold_12943 764427 94 + 5039921 ---------------CACGGCGCUC-----AUGACAAAUGGCCUGAAGUUAGAUGCUCGAGAGCGCAUUCCUCGGAGGCAACAAGGCAUCCACAUACUUAUGUAUAUUGACUCG ---------------....((((((-----.(((((..((((.....))))..)).))).)))))).......(((.((......)).)))..((((....))))......... ( -24.00, z-score = -0.49, R) >consensus _______CACGAGCACAUGACAA_______AUGACAAGUGCCGUGGAGUUGGAUGCGUGAAAACGCAUUUCUCGG_GGCGACAAGGAGUCCACAUACUUAAG___ACGAACUCG ...................................(((((..(((((....(((((((....)))))))(((.(.......).)))..))))).)))))............... (-14.70 = -15.87 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:16 2011