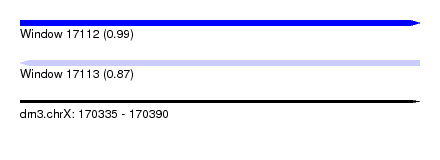
| Sequence ID | dm3.chrX |
|---|---|
| Location | 170,335 – 170,390 |
| Length | 55 |
| Max. P | 0.985039 |

| Location | 170,335 – 170,390 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 88.34 |
| Shannon entropy | 0.24460 |
| G+C content | 0.52873 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -13.42 |
| Energy contribution | -13.31 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
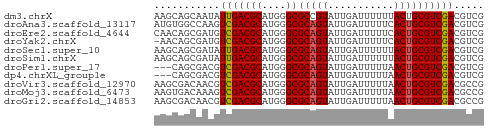
>dm3.chrX 170335 55 + 22422827 CGACGUCGACGCAGUAAAAAAUCAAUACGGCGCCCAUGCGUCAAUAUUGCUGCUU ..........(((((((.(......)..(((((....)))))....))))))).. ( -11.90, z-score = -0.17, R) >droAna3.scaffold_13117 4811313 55 + 5790199 CGACGUCGACGCAGUGAAAAAUCAAUACUGCGCCCAUGCGUCGACUUGGCCACAU (((.((((((((((((.........))))))((....)))))))))))....... ( -18.10, z-score = -2.34, R) >droEre2.scaffold_4644 117986 55 + 2521924 CGACGUCGACGCAGUGAAAAAUCAAUACUGCGCCCAUGCGUCGACAUCGCUGUUG (((.((((((((((((.........))))))((....)))))))).)))...... ( -19.90, z-score = -2.75, R) >droYak2.chrX 113436 54 + 21770863 CGACGUCGACGCAGUGAAAAAUCAAUACUGCGCCCAUGCGUCGACAUCGCUGUU- (((.((((((((((((.........))))))((....)))))))).))).....- ( -19.90, z-score = -2.87, R) >droSec1.super_10 42229 55 + 3036183 CGACGUCGACGCAGUAAAAAAUCAAUACUGCGCCCAUGCGUCAAUAUCGCUGCUU .(((((.(.(((((((.........))))))).)...)))))............. ( -15.10, z-score = -2.11, R) >droSim1.chrX 113704 55 + 17042790 CGACGUCGACGCAGUAAAAAAUCAAUACUGCGCCCAUGCGUCAAUAUCGCUGCUU .(((((.(.(((((((.........))))))).)...)))))............. ( -15.10, z-score = -2.11, R) >droPer1.super_17 1031240 52 + 1930428 CGACGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACGUCGCUG--- (((((((((((((((...........)))))((....))))))))))))...--- ( -25.40, z-score = -5.56, R) >dp4.chrXL_group1e 4089975 52 + 12523060 CGACGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACGUCGCUG--- (((((((((((((((...........)))))((....))))))))))))...--- ( -25.40, z-score = -5.56, R) >droVir3.scaffold_12970 10378410 55 + 11907090 CGGCGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACGUUGUCGCUU (((((((((((((((...........)))))((....))))))))))))...... ( -23.00, z-score = -3.46, R) >droMoj3.scaffold_6473 5863373 55 - 16943266 CGGCGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACUUUGUCACUU .((((((((((((((...........)))))((....))))))))...))).... ( -18.40, z-score = -2.87, R) >droGri2.scaffold_14853 3808164 55 - 10151454 CGGCGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACGUUGUCGCUU (((((((((((((((...........)))))((....))))))))))))...... ( -23.00, z-score = -3.46, R) >consensus CGACGUCGACGCAGUUAAAAAUCAAUACUGCGCCCAUGCGUCGACAUCGCUGCUU .(((((.(.((((((...........)))))).)...)))))............. (-13.42 = -13.31 + -0.11)

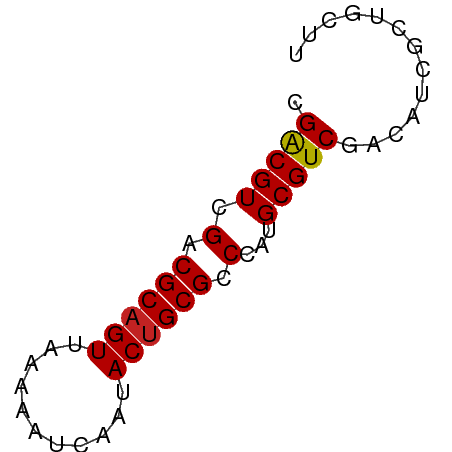
| Location | 170,335 – 170,390 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 88.34 |
| Shannon entropy | 0.24460 |
| G+C content | 0.52873 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -13.03 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 170335 55 - 22422827 AAGCAGCAAUAUUGACGCAUGGGCGCCGUAUUGAUUUUUUACUGCGUCGACGUCG .............((((..(.(((((.(((.........))).))))).))))). ( -14.00, z-score = -0.35, R) >droAna3.scaffold_13117 4811313 55 - 5790199 AUGUGGCCAAGUCGACGCAUGGGCGCAGUAUUGAUUUUUCACUGCGUCGACGUCG ....(((...((((((((....))(((((...((....)))))))))))))))). ( -18.70, z-score = -1.34, R) >droEre2.scaffold_4644 117986 55 - 2521924 CAACAGCGAUGUCGACGCAUGGGCGCAGUAUUGAUUUUUCACUGCGUCGACGUCG ......((((((((((((....))(((((...((....))))))))))))))))) ( -24.00, z-score = -3.59, R) >droYak2.chrX 113436 54 - 21770863 -AACAGCGAUGUCGACGCAUGGGCGCAGUAUUGAUUUUUCACUGCGUCGACGUCG -.....((((((((((((....))(((((...((....))))))))))))))))) ( -24.00, z-score = -3.61, R) >droSec1.super_10 42229 55 - 3036183 AAGCAGCGAUAUUGACGCAUGGGCGCAGUAUUGAUUUUUUACUGCGUCGACGUCG .............((((..(.(((((((((.........))))))))).))))). ( -18.60, z-score = -2.18, R) >droSim1.chrX 113704 55 - 17042790 AAGCAGCGAUAUUGACGCAUGGGCGCAGUAUUGAUUUUUUACUGCGUCGACGUCG .............((((..(.(((((((((.........))))))))).))))). ( -18.60, z-score = -2.18, R) >droPer1.super_17 1031240 52 - 1930428 ---CAGCGACGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGUCG ---...((((((((((((....))(((((...........))))))))))))))) ( -25.50, z-score = -4.48, R) >dp4.chrXL_group1e 4089975 52 - 12523060 ---CAGCGACGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGUCG ---...((((((((((((....))(((((...........))))))))))))))) ( -25.50, z-score = -4.48, R) >droVir3.scaffold_12970 10378410 55 - 11907090 AAGCGACAACGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGCCG .........(((((((((....))(((((...........))))))))))))... ( -19.20, z-score = -1.76, R) >droMoj3.scaffold_6473 5863373 55 + 16943266 AAGUGACAAAGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGCCG ..........((((((((....))(((((...........))))))))))).... ( -17.80, z-score = -1.68, R) >droGri2.scaffold_14853 3808164 55 + 10151454 AAGCGACAACGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGCCG .........(((((((((....))(((((...........))))))))))))... ( -19.20, z-score = -1.76, R) >consensus AAGCAGCGAUGUCGACGCAUGGGCGCAGUAUUGAUUUUUAACUGCGUCGACGUCG ...........(((((((....))(((((...........))))))))))..... (-13.03 = -12.92 + -0.11)
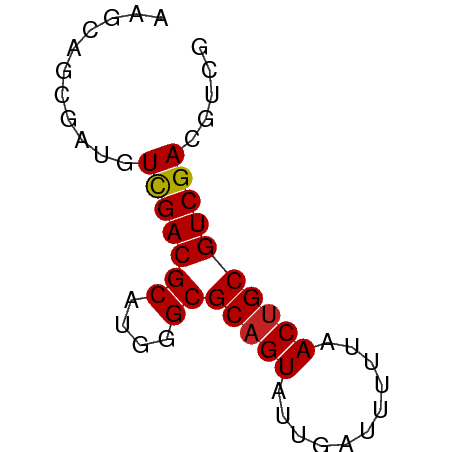
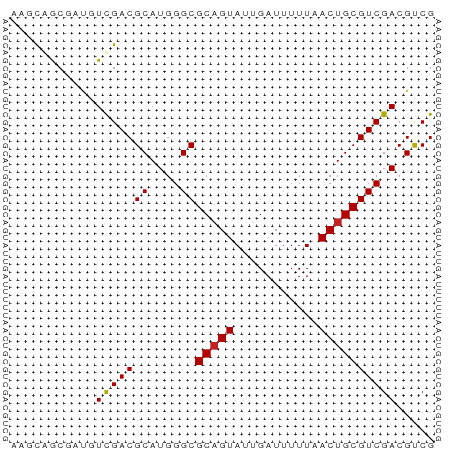
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:26 2011