
| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 203,355 – 203,543 |
| Length | 188 |
| Max. P | 0.996726 |

| Location | 203,355 – 203,448 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 63.06 |
| Shannon entropy | 0.62070 |
| G+C content | 0.57832 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
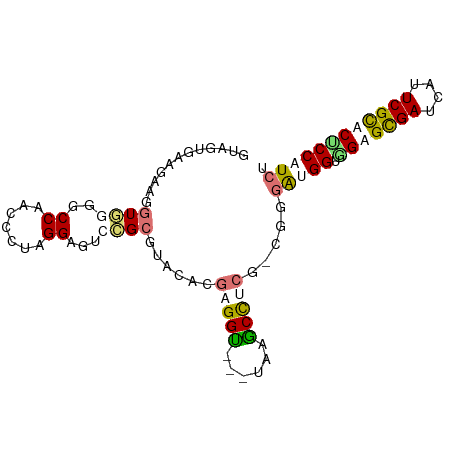
>dm3.chrXHet 203355 93 + 204112 GUGCGGACGAUGGUUGCACCCUCCCUCGGAGUCAGAGUACACGAGGUACGUGAGCUCCGUCGAGGUGGUGGUGCGAUCAUUCGUACUCCUUCG (((((...(((((((((((((..((((((.....((((.((((.....)))).))))..))))))..).)))))))))))))))))....... ( -44.30, z-score = -3.30, R) >dp4.Unknown_singleton_2130 3913 90 + 5907 GUUGUGAAAGAGGUGGGGCCAUCCUUGGGGGUGCGCAUACUUGAGGU---UAAGCCUCGCCGUGAUGGUAGUGUGAUUAUUCGCACUCCAUCU ...........((((((((...(((..((.(....)...))..))).---...))))))))..(((((.(((((((....)))))))))))). ( -40.80, z-score = -4.06, R) >droPer1.super_92 115677 90 - 190782 GUCAAGAAGACCGUGGUGCCGACACUUGGAGUCCGCGUCCACGAGGCUCGCGAGCUG---AAGAGCGGAGGAGCGAUUAUUCGCACCCCUUCA (((.....)))((((((((.(((.......))).))).)))))..((((........---..))))(((((.((((....))))...))))). ( -32.50, z-score = -0.75, R) >droGri2.scaffold_15056 30690 90 - 38174 GUAGUGAAGGAGGUAGGGCCUACCUUAGGUGUGCGCGUCCAUGAGGU---GAAACCUCUGCGGGAUGGUGGAGCGAUCAUUCGCACACCAUCU ((((..(((((((.....))).))))((((...(((.((...)).))---)..)))))))).((((((((..((((....)))).)))))))) ( -37.20, z-score = -2.44, R) >consensus GUAGUGAAGAAGGUGGGGCCAACCCUAGGAGUCCGCGUACACGAGGU___UAAGCCUCG_CGGGAUGGUGGAGCGAUCAUUCGCACUCCAUCU ............((((..((.......))...))))......(((((......))))).....(((((.(((((((....)))))))))))). (-16.20 = -17.70 + 1.50)

| Location | 203,390 – 203,488 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 63.08 |
| Shannon entropy | 0.60299 |
| G+C content | 0.52994 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
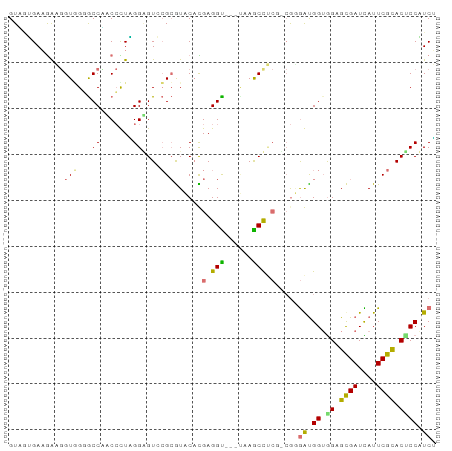
>dm3.chrXHet 203390 98 + 204112 AGUACACGAGGUACGUGAGCUCCGUCGAGGUGGUGGUGCGAUCAUUCGUACUCCUUCGGUUGGAGAGCUGCAGAAGGUGAUGGCAUCGAAAAGAUUCG ..(((.(((((..(....)..)).)))..))).((((((.((((((((((((((.......))))...))).))..))))).)))))).......... ( -27.60, z-score = 0.72, R) >dp4.Unknown_singleton_2130 3948 95 + 5907 CAUACUUGAGGU---UAAGCCUCGCCGUGAUGGUAGUGUGAUUAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAAAUUGCGGAAAAUGCAAAGUUUG ((.((((((((.---....)))).(((..((((.(((((((....))))))))).((((((....)))))).....))..)))........)))).)) ( -32.20, z-score = -3.17, R) >droPer1.super_92 115712 95 - 190782 CGUCCACGAGGCUCGCGAGCU---GAAGAGCGGAGGAGCGAUUAUUCGCACCCCUUCAGUGAGCGAACUGAGGAAGGUAGUGGCCAGCAGCAAAUUUA .(.((((...((((((..(((---....)))(((((.((((....))))...))))).))))))..(((......))).)))).)............. ( -31.20, z-score = -0.70, R) >droGri2.scaffold_15056 30725 95 - 38174 CGUCCAUGAGGU---GAAACCUCUGCGGGAUGGUGGAGCGAUCAUUCGCACACCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGGGAACGCAAAAUUCA ..(((..((((.---....)))).(((((((((((..((((....)))).))))))))..))).....))).(((..(((((.....)))))..))). ( -38.20, z-score = -2.80, R) >consensus CGUACACGAGGU___UAAGCCUCGGCGGGAUGGUGGAGCGAUCAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGCAAACACAAAAUUCA .......(((((......)))))........((.(((((((....))))))))).(((((......))))).(((..(((((.....)))))..))). (-20.10 = -20.22 + 0.12)



| Location | 203,412 – 203,528 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 57.16 |
| Shannon entropy | 0.70119 |
| G+C content | 0.51507 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
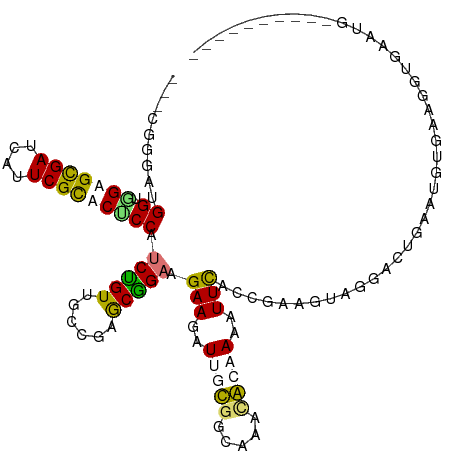
>dm3.chrXHet 203412 116 + 204112 CGUCGAGGUGGUGGUGCGAUCAUUCGUACUCCUUCGGUUGGAGAGCUGCAGAAGGUGAUGGCAUCGAAAAGAUUCGCAGAACUUGGACUGAAUGUGGCACGGAACGCGGCCGAGAA ..((..(((.(((((((((....))))))((((((((((.(((..((((....((((....))))(((....)))))))..))).))))))).(.....)))).))).)))..)). ( -41.50, z-score = -1.49, R) >dp4.Unknown_singleton_2130 3974 99 + 5907 -------AUGGUAGUGUGAUUAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAAAUUGCGGAAAAUGCAAAGUUUGAUGAGGUAGGACUGACUGUGAAGGUCAAUG---------- -------((((.(((((((....)))))))))))...(((((...((...(((..(((((.....)))))..)))..)).)))))...(((((.....)))))...---------- ( -28.40, z-score = -2.09, R) >droPer1.super_92 115734 103 - 190782 ---AAGAGCGGAGGAGCGAUUAUUCGCACCCCUUCAGUGAGCGAACUGAGGAAGGUAGUGGCCAGCAGCAAAUUUACCGAAGCAGGACUGGAGGUUAAGAAGAGUC---------- ---.........((.((((....)))).))((((((((.......(((.....(((((..((.....))....)))))....))).))))))))............---------- ( -26.00, z-score = -0.10, R) >droGri2.scaffold_15056 30747 113 - 38174 ---CGGGAUGGUGGAGCGAUCAUUCGCACACCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGGGAACGCAAAAUUCAGCGAAGUUGGUCUGGAAGUGAGUGUGAAAGAGAGACUGGG ---..((((((((..((((....)))).))))))))....((..((....(((..(((((.....)))))..))).))..((((..(((...............)))..)))))). ( -35.26, z-score = -1.51, R) >consensus ___CGGGAUGGUGGAGCGAUCAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGCAAACACAAAAUUCACCGAAGUAGGACUGAAUGUGAAGGUGAAUG__________ .........((.(((((((....))))))))).(((((......))))).(((..(((((.....)))))..)))......................................... (-16.38 = -16.38 + 0.00)


| Location | 203,424 – 203,543 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 53.38 |
| Shannon entropy | 0.73869 |
| G+C content | 0.50983 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.07 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
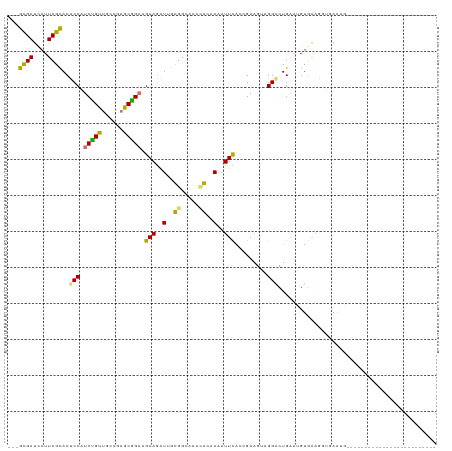
>dm3.chrXHet 203424 119 + 204112 GGUGCGAUCAUUCGUACUCCUUCGGUUGGAGAGCUGCAGAAGGUGAUGGCAUCGAAAAGAUUCGCAGAACUUGGACUGAAUGUGGCACGGAACGCGGCCGAGAAGCCGAAGGUCAUAGU (((((((....)))))))..(((((((.(((..((((....((((....))))(((....)))))))..))).)))))))((((((.(......((((......))))..))))))).. ( -40.70, z-score = -1.30, R) >dp4.Unknown_singleton_2130 3982 91 + 5907 ---GUGAUUAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAAAUUGCGGAAAAUGCAAAGUUUGAUGAGGUAGGACUGACUGUGAAGGUCAAUG------------------------- ---.(((((.((((((.((..(((..((((...((...(((..(((((.....)))))..)))..)).)))))))..)).)))))))))))...------------------------- ( -22.60, z-score = -0.82, R) >droPer1.super_92 115746 91 - 190782 ---GCGAUUAUUCGCACCCCUUCAGUGAGCGAACUGAGGAAGGUAGUGGCCAGCAGCAAAUUUACCGAAGCAGGACUGGAGGUUAAGAAGAGUC------------------------- ---((((....))))...((((((((.......(((.....(((((..((.....))....)))))....))).))))))))............------------------------- ( -23.80, z-score = -0.48, R) >droGri2.scaffold_15056 30759 116 - 38174 ---GCGAUCAUUCGCACACCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGGGAACGCAAAAUUCAGCGAAGUUGGUCUGGAAGUGAGUGUGAAAGAGAGACUGGGGCCGAAAGUGGUCGU ---((((((((((((......((((((....)))))).(((..(((((.....)))))..))).))))..(((((((...(((.....(....)....))).)))))))..)))))))) ( -36.40, z-score = -0.86, R) >consensus ___GCGAUCAUUCGCACUCCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGCAAACACAAAAUUCACCGAAGUAGGACUGAAUGUGAAGGUGAAUG_________________________ ...((((....))))..(((.(((((......))))).(((..(((((.....)))))..))).........)))............................................ (-13.57 = -13.07 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:19 2011