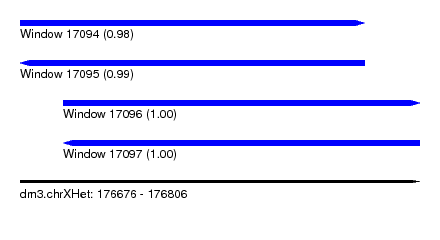
| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 176,676 – 176,806 |
| Length | 130 |
| Max. P | 0.998509 |

| Location | 176,676 – 176,788 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 61.19 |
| Shannon entropy | 0.63999 |
| G+C content | 0.50107 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -16.39 |
| Energy contribution | -18.45 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 176676 112 + 204112 -GUGGUCGAGCUGAGGCGUGCCUCAGCCAACUACAAGAAGCUCAUUGCGAGGGCGAAAAUGGAUGACUGGAAACGCUUCGUGGGAGAUCAUGCCGACGACCCACGGGGGCGCG -(((((.(.(((((((....)))))))).)))))......(((.....))).(((.............(....).(((((((((...((.....))...))))))))).))). ( -41.90, z-score = -1.13, R) >droAna3.scaffold_3813 5898 111 + 11518 GGGAUCAGCUUAGUACUAGGGUUC--AUGAGUACAAGCAUAGGGUUUUGAGAGUAAAAGAAGAUGAGUGGCGCUCUUUCGUAGAACGGUAUAAACGCGAUCCCUGGGGUCGUG ((((((.((((.(((((.......--...)))))))))...(.((((.....((....((((..(((.....))))))).....)).....)))).))))))).......... ( -27.20, z-score = 0.05, R) >droVir3.scaffold_12401 571 112 + 1125 -AGGAUUGAAUAUAGUCGGUGUAUGAAUGAGUACAAGCAAAUGCUUGUAAAGGUGAAAGAAGAUGAAUGGCGCUCUUUCCUGGAGCGCAAUAAGGAUGACCCCUGGGGUCGUG -.(.(((((......))))).).........(((((((....)))))))....................(((((((.....))))))).......(((((((...))))))). ( -31.80, z-score = -1.99, R) >droGri2.scaffold_15056 32355 112 - 38174 -AGGUUAGCUUACAGGCAGUGUAUGAAAGAGUACAAGCAAAUGCUUGUAAGAGUGAAAGAGAAUGAAUGGCGUUCCUUUUUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUG -.......((((((((((.(((((......)))))......))))))))))..................(((((((.....))))))).......(((((((...))))))). ( -40.00, z-score = -4.27, R) >consensus _AGGUUAGAUUAGAGGCAGGGUACGAAUGAGUACAAGCAAAUGCUUGUAAGAGUGAAAGAAGAUGAAUGGCGCUCCUUCGUGGAACGCAAUAAGGACGACCCCUGGGGUCGUG ...............................(((((((....))))))).................((((((((((.....))))))))))....(((((((...))))))). (-16.39 = -18.45 + 2.06)

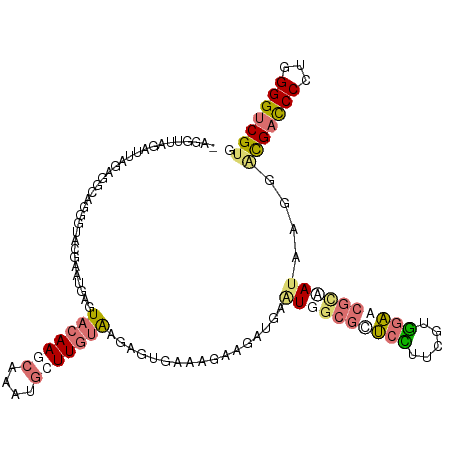
| Location | 176,676 – 176,788 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.19 |
| Shannon entropy | 0.63999 |
| G+C content | 0.50107 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -10.57 |
| Energy contribution | -12.95 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 176676 112 - 204112 CGCGCCCCCGUGGGUCGUCGGCAUGAUCUCCCACGAAGCGUUUCCAGUCAUCCAUUUUCGCCCUCGCAAUGAGCUUCUUGUAGUUGGCUGAGGCACGCCUCAGCUCGACCAC- .((((...((((((..(((.....)))..))))))..)))).....(((.................((((..((.....)).))))(((((((....)))))))..)))...- ( -36.10, z-score = -0.95, R) >droAna3.scaffold_3813 5898 111 - 11518 CACGACCCCAGGGAUCGCGUUUAUACCGUUCUACGAAAGAGCGCCACUCAUCUUCUUUUACUCUCAAAACCCUAUGCUUGUACUCAU--GAACCCUAGUACUAAGCUGAUCCC ..........((((((((((((....((.....))...))))))...............................(((((((((...--.......))))).)))).)))))) ( -23.60, z-score = -2.48, R) >droVir3.scaffold_12401 571 112 - 1125 CACGACCCCAGGGGUCAUCCUUAUUGCGCUCCAGGAAAGAGCGCCAUUCAUCUUCUUUCACCUUUACAAGCAUUUGCUUGUACUCAUUCAUACACCGACUAUAUUCAAUCCU- ...(((((...))))).........((((((.......))))))....................(((((((....)))))))..............................- ( -25.50, z-score = -3.24, R) >droGri2.scaffold_15056 32355 112 + 38174 CACGACCCCAGGGGUCGUCCUUAUUGCGCUCCAAAAAGGAACGCCAUUCAUUCUCUUUCACUCUUACAAGCAUUUGCUUGUACUCUUUCAUACACUGCCUGUAAGCUAACCU- .(((((((...))))))).(((((.(((.(((.....))).)))....................(((((((....)))))))..................))))).......- ( -27.10, z-score = -3.20, R) >consensus CACGACCCCAGGGGUCGUCCUUAUUGCGCUCCACGAAAGAGCGCCAUUCAUCCUCUUUCACCCUCACAAGCAUUUGCUUGUACUCAUUCAAACACCGCCUCUAAGCUAACCC_ .(((((((...))))))).......(((((((.....))))))).....................((((((....))))))................................ (-10.57 = -12.95 + 2.38)

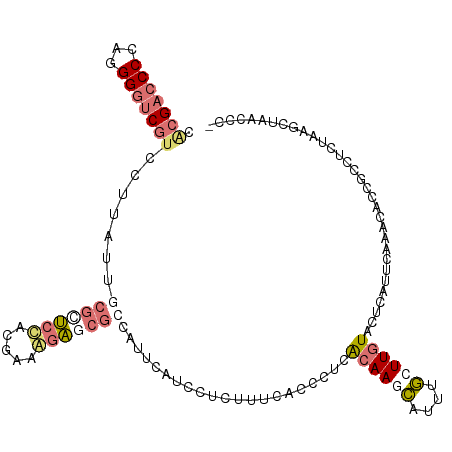
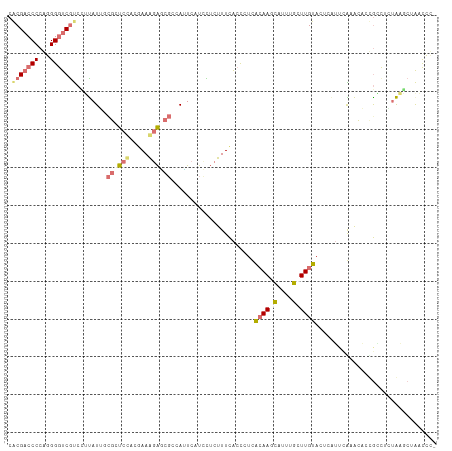
| Location | 176,690 – 176,806 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.20 |
| Shannon entropy | 0.56846 |
| G+C content | 0.47699 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -17.83 |
| Energy contribution | -19.82 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 176690 116 + 204112 GCGUGCCUCAGCCAACUACAAGAAGCUCAUUGCGAGGGCGAAAAUGGAUGACUGGAAACGCUUCGUGGGAGAUCAUGCCGACGACCCACGGGGGCGCGUCUACAAGAUUUGCCGAG (((((((((............(((((...((((....))))............(....))))))(((((...((.....))...))))).)))))))))................. ( -36.10, z-score = -0.01, R) >droAna3.scaffold_3813 5915 112 + 11518 ---AGGGUUCAUGAG-UACAAGCAUAGGGUUUUGAGAGUAAAAGAAGAUGAGUGGCGCUCUUUCGUAGAACGGUAUAAACGCGAUCCCUGGGGUCGUGUUUACAAAAUUCUGCGGG ---.........(((-(.((..(((....((((........))))..)))..))..))))..((((((((.....((((((((((((...)))))))))))).....)))))))). ( -32.90, z-score = -1.89, R) >droVir3.scaffold_12401 589 112 + 1125 ---UGUAUGAAUGAG-UACAAGCAAAUGCUUGUAAAGGUGAAAGAAGAUGAAUGGCGCUCUUUCCUGGAGCGCAAUAAGGAUGACCCCUGGGGUCGUGUUUAUAAAAUAGUUCGAG ---............-(((((((....)))))))..............(((((.(((((((.....))))))).(((((.(((((((...))))))).)))))......))))).. ( -37.10, z-score = -4.25, R) >droGri2.scaffold_15056 32373 112 - 38174 ---UGUAUGAAAGAG-UACAAGCAAAUGCUUGUAAGAGUGAAAGAGAAUGAAUGGCGUUCCUUUUUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUGCUUAUAAAGUUGUGAGAG ---............-(((((((....)))))))....................(((((((.....))))))).(((((.(((((((...))))))).)))))............. ( -38.40, z-score = -5.11, R) >consensus ___UGUAUGAAUGAG_UACAAGCAAAUGCUUGUAAGAGUGAAAGAAGAUGAAUGGCGCUCCUUCGUGGAACGCAAUAAGGACGACCCCUGGGGUCGUGUUUACAAAAUUGUGCGAG ................(((((((....)))))))..................(((((((((.....)))))))))((((.(((((((...))))))).)))).............. (-17.83 = -19.82 + 2.00)


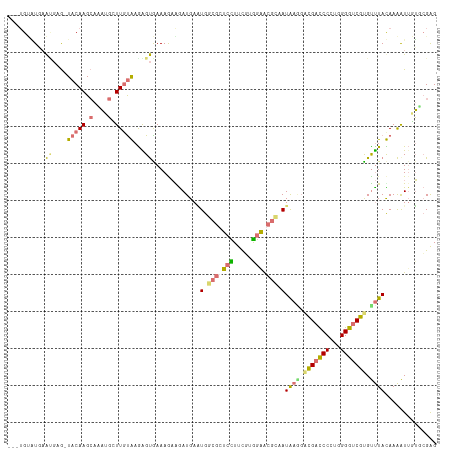
| Location | 176,690 – 176,806 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.20 |
| Shannon entropy | 0.56846 |
| G+C content | 0.47699 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -12.35 |
| Energy contribution | -15.35 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 176690 116 - 204112 CUCGGCAAAUCUUGUAGACGCGCCCCCGUGGGUCGUCGGCAUGAUCUCCCACGAAGCGUUUCCAGUCAUCCAUUUUCGCCCUCGCAAUGAGCUUCUUGUAGUUGGCUGAGGCACGC ((((((.....(((.((((((.....((((((..(((.....)))..))))))..)))))).)))...................((((..((.....)).))))))))))...... ( -32.70, z-score = 0.58, R) >droAna3.scaffold_3813 5915 112 - 11518 CCCGCAGAAUUUUGUAAACACGACCCCAGGGAUCGCGUUUAUACCGUUCUACGAAAGAGCGCCACUCAUCUUCUUUUACUCUCAAAACCCUAUGCUUGUA-CUCAUGAACCCU--- ..((((((((..(((((((.(((((....)).))).)))))))..))))).(....).))).......................................-............--- ( -21.70, z-score = -1.94, R) >droVir3.scaffold_12401 589 112 - 1125 CUCGAACUAUUUUAUAAACACGACCCCAGGGGUCAUCCUUAUUGCGCUCCAGGAAAGAGCGCCAUUCAUCUUCUUUCACCUUUACAAGCAUUUGCUUGUA-CUCAUUCAUACA--- ...(((...............(((((...))))).........((((((.......))))))..)))...............(((((((....)))))))-............--- ( -26.50, z-score = -3.66, R) >droGri2.scaffold_15056 32373 112 + 38174 CUCUCACAACUUUAUAAGCACGACCCCAGGGGUCGUCCUUAUUGCGCUCCAAAAAGGAACGCCAUUCAUUCUCUUUCACUCUUACAAGCAUUUGCUUGUA-CUCUUUCAUACA--- .............(((((.(((((((...))))))).))))).(((.(((.....))).)))....................(((((((....)))))))-............--- ( -29.70, z-score = -5.74, R) >consensus CUCGCACAAUUUUAUAAACACGACCCCAGGGGUCGUCCUUAUUGCGCUCCACGAAAGAGCGCCAUUCAUCCUCUUUCACCCUCACAAGCAUUUGCUUGUA_CUCAUUCAUACA___ .............(((((.(((((((...))))))).))))).(((((((.....))))))).....................((((((....))))))................. (-12.35 = -15.35 + 3.00)

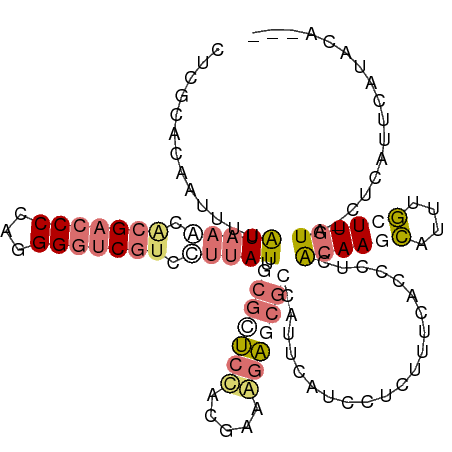
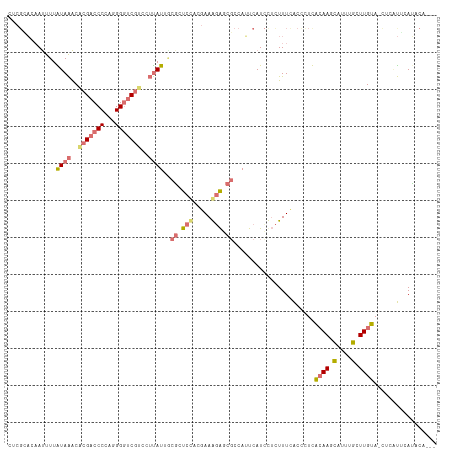
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:13 2011