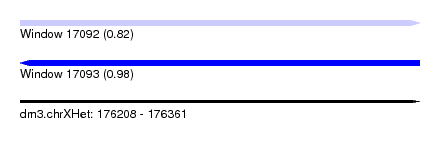
| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 176,208 – 176,361 |
| Length | 153 |
| Max. P | 0.980405 |

| Location | 176,208 – 176,361 |
|---|---|
| Length | 153 |
| Sequences | 4 |
| Columns | 160 |
| Reading direction | forward |
| Mean pairwise identity | 52.87 |
| Shannon entropy | 0.78589 |
| G+C content | 0.48764 |
| Mean single sequence MFE | -51.51 |
| Consensus MFE | -18.51 |
| Energy contribution | -20.26 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
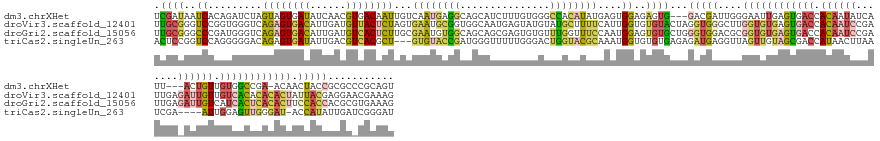
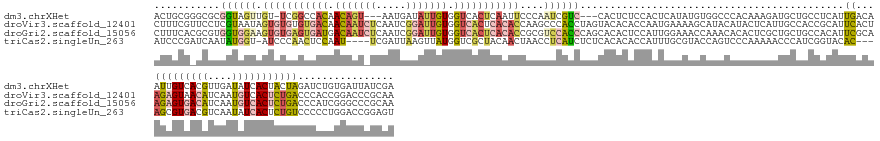
>dm3.chrXHet 176208 153 + 204112 UCGAUAAUCACAGAUCUAGUAGUGAUAUCAACGUGACAAUUGUCAAUGAGGCAGCAUCUUUGUGGGCCACAUAUGAGUGGAGAGUG---GACGAUUGGGAAUUGAGUGACCACAAUAUCAUU---ACUGUUGUGGCCGA-ACAACUACCGCGCCCGCAGU ..((((.((((..........))))))))....((((....))))(((.(((.(((....)))..))).)))....((((.(.(((---(.((((.....))))..((.((((((((.....---..)))))))).)).-.......)))).)))))... ( -40.00, z-score = 0.44, R) >droVir3.scaffold_12401 104 160 + 1125 UUGCGGGUCCGGUGGGUCAGAGUGACAUUGAUGUUACUCUAGUGAAUGCGGUGGCAAUGAGUAUGUAUGCUUUUCAUUGGUGUGUACUAGGUGGGCUUGGUGUGAGUGACCACAAUCCGAUUGAGAUUGUUGUCACACACACUAUUACGAGGAACGAAAG ...((((((((.(.(((((((((((((....))))))))(((((((.(((...(((.......))).)))..)))))))...)).))).).))))))))(((((.(((((.((((((.......)))))).))))).)))))............(....) ( -53.00, z-score = -1.71, R) >droGri2.scaffold_15056 31888 160 - 38174 UUGCGGGCCCGAUGGGUCAGAGUGACAUUGAUGUCACUCUUGCGAAUGUGGCAGCAGCGAGUGUGUUUGGUUUCCAAUGGAGUGUGCUGGGUGGACGCGGUGUGAGUGACCACAAUCCGAUUGAGAUUGUCAUCACUCACACUUCCACCACGCGUGAAAG ((((((((((...)))))(((((((((....))))))))))))))(((((....(((((.......((((...)))).......)))))((((((...(((((((((((..((((((.......))))))..))))))))))))))))).)))))..... ( -69.54, z-score = -3.28, R) >triCas2.singleUn_263 2728 152 - 6727 ACUCCGGUCCAGGGGGACAGAGUGAUAUUGACGUCACGCU---GUGUACCGAUGGGUUUUUGGGACUGGUACGCAAAUGGUGUGUGAGAGAUGAGGUUAGUUGUAGCGACCAUAACUUAAUCGA----AUUGGAGUUGGGAU-ACCAUAUUGAUCGGGAU ..(((((((..((.(.((((.(((((......))))).))---)).).)).((((..(((..(..(..(((((((.....)))))....(((.((((((((((...))))..)))))).)))..----))..)..)..))).-.))))...))))))).. ( -43.50, z-score = -0.27, R) >consensus UCGCGGGUCCGAGGGGUCAGAGUGACAUUGACGUCACUCUUGUGAAUGCGGCAGCAUCGAGGGGGUUUGCAUUCCAAUGGAGUGUGCUAGAUGAGCGUGGUGUGAGUGACCACAAUCCGAUUGA_AUUGUUGUCACUCACACUACCACCACGAACGAAAG ..((..(((.........((((((((......))))))))...((((((((...............))))))))...............)))..))...(((((((((((.((((((.......)))))).))))))))))).................. (-18.51 = -20.26 + 1.75)
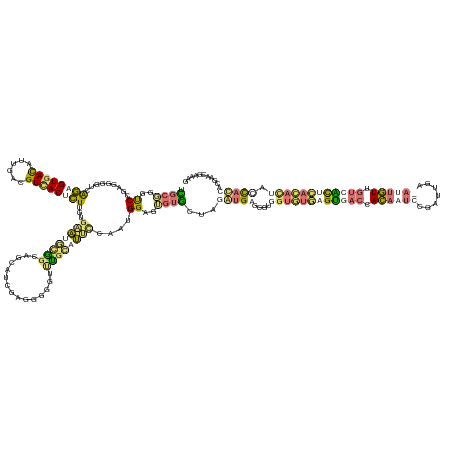
| Location | 176,208 – 176,361 |
|---|---|
| Length | 153 |
| Sequences | 4 |
| Columns | 160 |
| Reading direction | reverse |
| Mean pairwise identity | 52.87 |
| Shannon entropy | 0.78589 |
| G+C content | 0.48764 |
| Mean single sequence MFE | -43.96 |
| Consensus MFE | -12.90 |
| Energy contribution | -17.53 |
| Covariance contribution | 4.63 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 176208 153 - 204112 ACUGCGGGCGCGGUAGUUGU-UCGGCCACAACAGU---AAUGAUAUUGUGGUCACUCAAUUCCCAAUCGUC---CACUCUCCACUCAUAUGUGGCCCACAAAGAUGCUGCCUCAUUGACAAUUGUCACGUUGAUAUCACUACUAGAUCUGUGAUUAUCGA .....(((.((((((.((((-..(((((((.((..---..)).....((((.(...............).)---)))............))))))).))))...)))))))))..((((....))))..((((((((((..........)))).)))))) ( -43.06, z-score = -1.34, R) >droVir3.scaffold_12401 104 160 - 1125 CUUUCGUUCCUCGUAAUAGUGUGUGUGACAACAAUCUCAAUCGGAUUGUGGUCACUCACACCAAGCCCACCUAGUACACACCAAUGAAAAGCAUACAUACUCAUUGCCACCGCAUUCACUAGAGUAACAUCAAUGUCACUCUGACCCACCGGACCCGCAA ..................(((((.(((((.(((((((.....))))))).))))).)))))...((...............((((((.............))))))...(((.......((((((.(((....))).))))))......)))....)).. ( -38.24, z-score = -2.17, R) >droGri2.scaffold_15056 31888 160 + 38174 CUUUCACGCGUGGUGGAAGUGUGAGUGAUGACAAUCUCAAUCGGAUUGUGGUCACUCACACCGCGUCCACCCAGCACACUCCAUUGGAAACCAAACACACUCGCUGCUGCCACAUUCGCAAGAGUGACAUCAAUGUCACUCUGACCCAUCGGGCCCGCAA .......(((.((((((.(((((((((((.(((((((.....))))))).)))))))))))....))))))((((........((((...))))........))))..(((.........(((((((((....))))))))).........))).))).. ( -65.16, z-score = -4.82, R) >triCas2.singleUn_263 2728 152 + 6727 AUCCCGAUCAAUAUGGU-AUCCCAACUCCAAU----UCGAUUAAGUUAUGGUCGCUACAACUAACCUCAUCUCUCACACACCAUUUGCGUACCAGUCCCAAAAACCCAUCGGUACAC---AGCGUGACGUCAAUAUCACUCUGUCCCCCUGGACCGGAGU .............(((.-...)))(((((...----............(((((((...............................))).))))((((.....(((....)))..((---((.((((........)))).))))......)))).))))) ( -29.38, z-score = -0.66, R) >consensus AUUUCGAGCGUGGUAGUAGUGUCAGCGACAACAAU_UCAAUCAGAUUGUGGUCACUCAAACCAAGCCCACCUAGCACACACCAUUGAAAAGCAAACACAAAAAAUGCUGCCGCAUUCACAAGAGUGACAUCAAUAUCACUCUGACCCACCGGACCCGCAA .........((((.....(((((((((((.(((((((.....))))))).))))))))))).....))))..................................................(((((((((....))))))))).................. (-12.90 = -17.53 + 4.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:10 2011