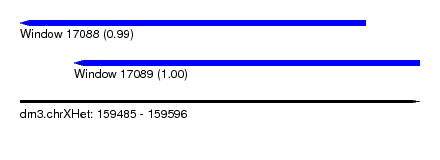
| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 159,485 – 159,596 |
| Length | 111 |
| Max. P | 0.999634 |

| Location | 159,485 – 159,581 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 51.77 |
| Shannon entropy | 0.64923 |
| G+C content | 0.56749 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -19.97 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
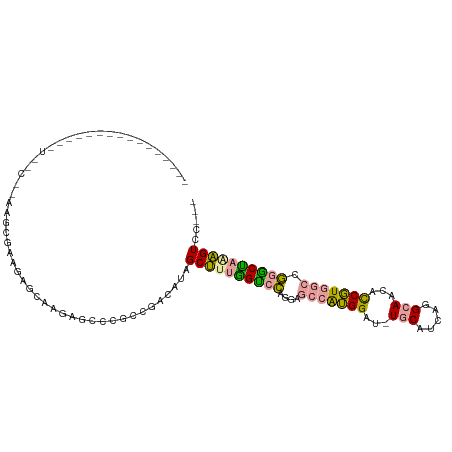
>dm3.chrXHet 159485 96 - 204112 ----------AUCACCAUCGCGCCAAGGCGGCCUUGGCCGUGCUGGCAAGGAUGUUCACCAAGCAACAUCUUGGGCUAGCCCUGGUGCAGGAACCCUGGUAUAACC-- ----------...(((...((((((.((((((....)))))((((((.((((((((........))))))))..))))))).))))))(((...))))))......-- ( -40.90, z-score = -2.02, R) >droYak2.chr3R 144096 90 - 28832112 ----------AGCAAGGGUCCGCCGACAUAGCUUUGGUCCAAGAGCCAUGGAU-UGCCUCGGGCAAUGCCGUCGCCGGACUAAAGUCCUCCAAUUA--CAACC----- ----------.((...((((((.((((...(((((......)))))...((((-((((...)))))).)))))).))))))...))..........--.....----- ( -30.30, z-score = -1.49, R) >dp4.Unknown_singleton_3065 1778 107 - 2449 AUUACCAUGGAGAAAGAGCCCGCCGACAUAGCUUUAGUCCAGGAGCCGUGGAU-UGCGUCAGGCAACUCCGUGGCCGGGCUAGAGUCCUCAAAUUAAGCAGUAAGCAG .......((..((...(((((((((((.........)))..)).(((((((((-(((.....)))).))))))))))))))....))..))......((.....)).. ( -32.60, z-score = -0.79, R) >consensus __________AGCAAGAGCCCGCCGACAUAGCUUUGGUCCAGGAGCCAUGGAU_UGCAUCAGGCAACACCGUGGCCGGGCUAAAGUCCUCAAAUUA_GCAAUAA_C__ ..............................((((((((((....(((((((...(((.....)))...))))))).))))))))))...................... (-19.97 = -19.53 + -0.43)


| Location | 159,500 – 159,596 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 47.95 |
| Shannon entropy | 0.70308 |
| G+C content | 0.58446 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -19.97 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrXHet 159500 96 - 204112 ---------------AUAGCUCAGGUGAAAAUCACCAUCGCGCCAAGGCGGCCUUGGCCGUGCUGGCAAGGAUGUUCACCAAGCAACAUCUUGGGCUAGCCCUGGUGCAGG ---------------........((((.....))))...((((((.((((((....)))))((((((.((((((((........))))))))..))))))).))))))... ( -43.60, z-score = -2.88, R) >droYak2.chr3R 144109 85 - 28832112 ----------------------AAGCGAAGAGCAAGGGUCCGCCGACAUAGCUUUGGUCCAAGAGCCAUGGAU-UGCCUCGGGCAAUGCCGUCGCCGGACUAAAGUCC--- ----------------------..((.....))...((((((.((((...(((((......)))))...((((-((((...)))))).)))))).)))))).......--- ( -32.00, z-score = -1.70, R) >dp4.Unknown_singleton_3065 1795 110 - 2449 UAGCGUCGGUGGAGCUUCUCAUUACCAUGGAGAAAGAGCCCGCCGACAUAGCUUUAGUCCAGGAGCCGUGGAU-UGCGUCAGGCAACUCCGUGGCCGGGCUAGAGUCCUCA ....((((((((.(((((((.........))))...)))))))))))...((((((((((..(.(((((((((-(((.....)))).)))))))))))))))))))..... ( -49.60, z-score = -3.50, R) >consensus ________________U__C__AAGCGAAGAGCAAGAGCCCGCCGACAUAGCUUUGGUCCAGGAGCCAUGGAU_UGCAUCAGGCAACACCGUGGCCGGGCUAAAGUCC___ ..................................................((((((((((....(((((((...(((.....)))...))))))).))))))))))..... (-19.97 = -19.53 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:06 2011