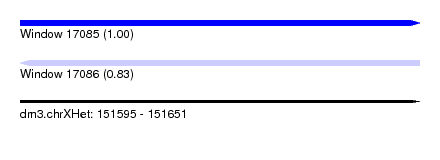
| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 151,595 – 151,651 |
| Length | 56 |
| Max. P | 0.995159 |

| Location | 151,595 – 151,651 |
|---|---|
| Length | 56 |
| Sequences | 3 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 66.07 |
| Shannon entropy | 0.47105 |
| G+C content | 0.40807 |
| Mean single sequence MFE | -10.27 |
| Consensus MFE | -10.52 |
| Energy contribution | -8.97 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.83 |
| Mean z-score | -2.35 |
| Structure conservation index | 1.02 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
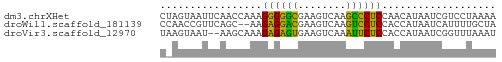
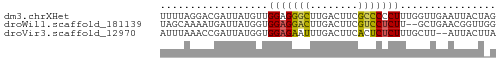
>dm3.chrXHet 151595 56 + 204112 CUAGUAAUUCAACCAAAGGGGGCGAAGUCAAGCCCUCCAACAUAAUCGUCCUAAAA .................((((((........)))).)).................. ( -11.30, z-score = -1.68, R) >droWil1.scaffold_181139 259246 54 - 724996 CCAACCGUUCAGC--AAGAGGACGAAGUCAAGUCCUCCACCAUAAUCAUUUUGCUA ..........(((--((((((((........)))))).............))))). ( -12.31, z-score = -3.19, R) >droVir3.scaffold_12970 10872781 54 - 11907090 UAAGUAAU--AAGCAAAGAGAGUGAAGUCAAAUUCUCCACCAUAAUCGGUUUAAAU ........--.......((((((........)))))).(((......)))...... ( -7.20, z-score = -2.19, R) >consensus CAAGUAAUUCAACCAAAGAGGGCGAAGUCAAGUCCUCCACCAUAAUCGUUUUAAAA .................((((((........))))))................... (-10.52 = -8.97 + -1.55)
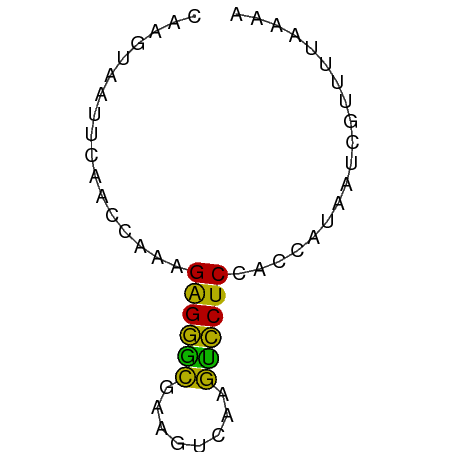
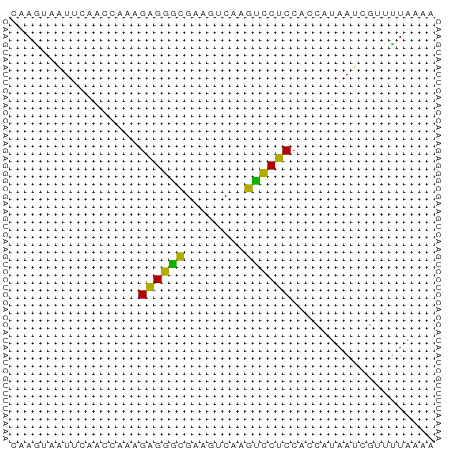
| Location | 151,595 – 151,651 |
|---|---|
| Length | 56 |
| Sequences | 3 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 66.07 |
| Shannon entropy | 0.47105 |
| G+C content | 0.40807 |
| Mean single sequence MFE | -11.24 |
| Consensus MFE | -7.47 |
| Energy contribution | -7.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
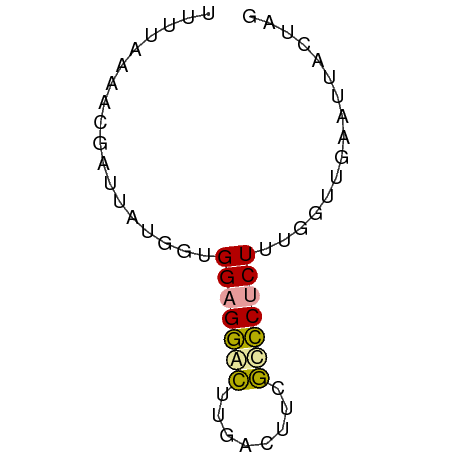
>dm3.chrXHet 151595 56 - 204112 UUUUAGGACGAUUAUGUUGGAGGGCUUGACUUCGCCCCCUUUGGUUGAAUUACUAG ........((((((....((.((((........))))))..))))))......... ( -13.50, z-score = -1.12, R) >droWil1.scaffold_181139 259246 54 + 724996 UAGCAAAAUGAUUAUGGUGGAGGACUUGACUUCGUCCUCUU--GCUGAACGGUUGG (((((.............(((((((........))))))))--))))......... ( -13.01, z-score = -1.17, R) >droVir3.scaffold_12970 10872781 54 + 11907090 AUUUAAACCGAUUAUGGUGGAGAAUUUGACUUCACUCUCUUUGCUU--AUUACUUA .........(((...((..((((...((....))...))))..)).--)))..... ( -7.20, z-score = -1.27, R) >consensus UUUUAAAACGAUUAUGGUGGAGGACUUGACUUCGCCCUCUUUGGUUGAAUUACUAG ..................(((((((........)))))))................ ( -7.47 = -7.03 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:03 2011