| Sequence ID | dm3.chrXHet |
|---|---|
| Location | 3,902 – 3,965 |
| Length | 63 |
| Max. P | 0.639626 |

| Location | 3,902 – 3,965 |
|---|---|
| Length | 63 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 59.56 |
| Shannon entropy | 0.81302 |
| G+C content | 0.43692 |
| Mean single sequence MFE | -14.84 |
| Consensus MFE | -4.50 |
| Energy contribution | -3.70 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
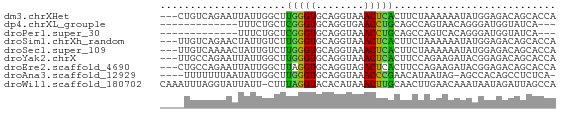
>dm3.chrXHet 3902 63 + 204112 ---CUGUCAGAAUUAUUGGCUUGGGUGCAGGUAAACUCACUUCUAAAAAAUAUGGAGACAGCACCA ---..(((((.....)))))...(((((((((......))))..........((....))))))). ( -14.30, z-score = -1.02, R) >dp4.chrXL_group1e 11967409 50 - 12523060 -------------UUUCUGCUCGGGUGCAGGUGAACCUGCAGCCAGUAACAGGGAUGGUAUCA--- -------------(..(((((.((.((((((....)))))).))))...)))..)........--- ( -20.10, z-score = -2.40, R) >droPer1.super_30 658354 50 + 958394 -------------UUUCUGCUCGGGUGCAGGUAAACCUGCAGCCAGUCACAGGGAUGGUAUCA--- -------------(..(((((.((.((((((....)))))).))))...)))..)........--- ( -18.60, z-score = -2.12, R) >droSim1.chrXh_random 3675 63 - 84659 ---UUGUCAGAACUAUUGUCUUGGGUGCAGGUAAACUCACUUCUAAAAAAUAUGGAGACAGCACCA ---....((..((....))..))(((((((((......))))..........((....))))))). ( -13.40, z-score = -0.76, R) >droSec1.super_109 3858 63 - 87397 ---UUGUCAAAACUAUUGUCUUGGGUGCAGGUAAACUCACUUCUAAAAAAUAUGGAGACAGCACCA ---....(((.((....)).)))(((((((((......))))..........((....))))))). ( -13.50, z-score = -1.27, R) >droYak2.chrX 21749578 63 + 21770863 ---UUGCCAGAAUUAUUGGCUUGGGUGCAGGUAAACUCACUUCCAGAAGAUACGGAGACAGCACCA ---..(((((.....)))))...(((((...........(((....)))....(....).))))). ( -16.80, z-score = -1.33, R) >droEre2.scaffold_4690 18681185 63 + 18748788 ---CUGCCAGAAUUAUUGGCUUAGGUGCAGGUAGACUCACUUCCAGAAGAUACGGAGACAGCACCA ---..(((((.....)))))...(((((.((.((.....)).)).........(....).))))). ( -18.50, z-score = -2.02, R) >droAna3.scaffold_12929 2268121 60 - 3277472 ----UUUUUUUAAUAUUGGCUUGGGUGCAGGUAAACCCGAACAUAAUAG-AGCCACAGCCUCUCA- ----............((((((((((........))))..........)-)))))..........- ( -11.60, z-score = -0.13, R) >droWil1.scaffold_180702 3715215 65 - 4511350 CAAAUUUAGGUAUUAUU-CUUUAGGUACACAUAAACUUGCAACUUGAACAAAUAAUAGAUUAGCCA ........(((((((((-.(((((((...((......))..)))))))..))))))......))). ( -6.80, z-score = -0.29, R) >consensus ____UGUCAGAAUUAUUGGCUUGGGUGCAGGUAAACUCACUUCUAGAAAAUAUGGAGACAGCACCA .....................(((((........)))))........................... ( -4.50 = -3.70 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:02:00 2011