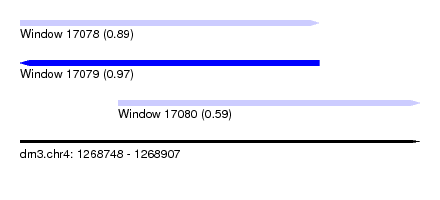
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 1,268,748 – 1,268,907 |
| Length | 159 |
| Max. P | 0.973697 |

| Location | 1,268,748 – 1,268,867 |
|---|---|
| Length | 119 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.43407 |
| G+C content | 0.41197 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -18.62 |
| Energy contribution | -19.59 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
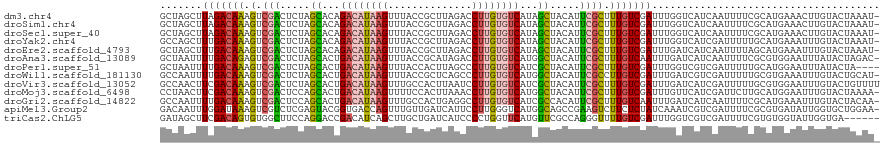
>dm3.chr4 1268748 119 + 1351857 -AUUUAGUACAAGUUUCAUGCGAAAAUUGAUGACCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGC -..(((((.(((.((((....)))).)))...........((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...))))). ( -30.60, z-score = -1.35, R) >droSim1.chr4 947137 119 + 949497 -AUUUAGUACAAGUUUCAUGCGAAAAUUGAUGACCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGC -..(((((.(((.((((....)))).)))...........((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...))))). ( -30.60, z-score = -1.35, R) >droSec1.super_40 286808 119 + 298270 -AUUUAGUACAAGUUUCAUGCGAAAAUUGAUGACCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGC -..(((((.(((.((((....)))).)))...........((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...))))). ( -30.60, z-score = -1.35, R) >droYak2.chr4 1370813 119 + 1374474 -AUUUAGUACAAAUUUCAUGCAAAAAUCGAUGACCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCAAAGCUGGC -..............(((.((.......(((......)))((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...))))). ( -29.80, z-score = -1.57, R) >droEre2.scaffold_4793 22122 119 - 55562 -AUUUAGUACAAAUUUCAUGCUAAAAUUGAUGAUCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCAAAGCUAGC -.(((((((.........)))))))...(((......)))((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))......... ( -29.70, z-score = -1.64, R) >droAna3.scaffold_13089 475457 119 + 978470 -GUCUAGUAUAAAUUCCACGCGAAAAUUGAUGAUCAAAUUGACAAAGCGAAUGUAGCCAUGACACAAGGUCUAUGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUCUGUCAAAAUUAGC -..(((((..................(((.....))).((((((.(((((.(.((((..(((((.(((..((....))....))).)))))..)))).).))).)).)))))).))))). ( -26.40, z-score = -0.83, R) >droPer1.super_51 489175 116 + 524598 ----UAGUAUAAAUUCCAUGCAAAAAUCGACGACCAAAUCGACAAAGCGAAUGUAGCGAUGACACAAGGGCUAAGUGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUAGC ----..((((.......))))...................((((((((((.(.(((((.(((((.(((.(((....)))...))).))))).))))).).))).)))))))......... ( -31.50, z-score = -3.07, R) >droWil1.scaffold_181130 16136723 119 - 16660200 -AUGCAGUACAAAUUUCACGCAAAAAUCGACGAUCAAAUCGACAAGGCGAAUGUAGCCAUGACACAAGGGCUGAGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUGGC -.(((.((.........)))))..................((((((((((.(.((((..(((((.(((.(((....)))...))).)))))..)))).).))).)))))))......... ( -30.30, z-score = -1.06, R) >droVir3.scaffold_13052 604576 120 + 2019633 AAAACAGUACAAAUUCCACGCAAAAAUCGAUGAUCAAAUCGACAAAGCGAAUGUAGCGAUGACACAAGGAUUAAGUGGCAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCGAAGUUGGC .................................((((.((((((((((((.(.(((((.(((((.(((..............))).))))).))))).).))).)))))))))..)))). ( -32.34, z-score = -2.42, R) >droMoj3.scaffold_6498 1841579 119 - 3408170 -UUUUAGUACAAAUUCCAUGCAAGAAUCGAUGAACAAAUCGACAAAGCGAAUGUAGCCAUGACACAAGGUUUAAGUGGAAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCGAAGUUAGG -.....((.((.((((.......))))...)).))...((((((((((((.(.((((..(((((..((((((.......)))))).)))))..)))).).))).)))))))))....... ( -30.80, z-score = -2.08, R) >droGri2.scaffold_14822 824072 119 - 1097346 -UUGUAGUACAAAUUUCAUGCGAAAAUUGAUGAUCAAAUUGACAAAGCGAAUGUGGCGAUGACACAAGGCCUCAGUGGCAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCAAAAUUGGC -(((..((.(((.((((....)))).)))))...))).((((((((((((.(.(((((.(((((.((((((.....)))...))).))))).))))).).))).)))))))))....... ( -35.70, z-score = -2.42, R) >apiMel3.Group2 14377538 119 - 14465785 -UUCCAGCACCAAUAUCACGCGAAAAUCGACGAUUUGAUAGAGAAGACUUCGGCUGCCAUGACCCAAGGAAUGAUCAACAAACUGGUCACCGUACUCGAGACGACUUUAUCCAAAUUGUC -.....((...........)).......(((((((((((((((..(..((((..(((..(((((..((...((.....))..)))))))..)))..)))).)..)))))).))))))))) ( -26.30, z-score = -1.16, R) >triCas2.ChLG5 10946359 114 - 18847211 ------UCACCAAUACCACACGAAAAUCGACGACCAAAUCGACAAAACCCUGGCGAACAUGAACCAGGGGAUGAUCAGCAAGCUGAUGUCGGUCCUGGAAGCCACACUGUCGAAGCUAUC ------....................(((((..(((.((((((....((((((..........))))))....(((((....)))))))))))..))).((.....)))))))....... ( -31.60, z-score = -2.06, R) >consensus _AUUUAGUACAAAUUUCAUGCGAAAAUCGAUGACCAAAUCGACAAAGCGAAUGUAGCCAUGACACAAGGUCUAAGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAGUUAGC ........................................((((((((((.(.((((...((((.(((..............))).))))...)))).).))).)))))))......... (-18.62 = -19.59 + 0.97)
| Location | 1,268,748 – 1,268,867 |
|---|---|
| Length | 119 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.43407 |
| G+C content | 0.41197 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.31 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
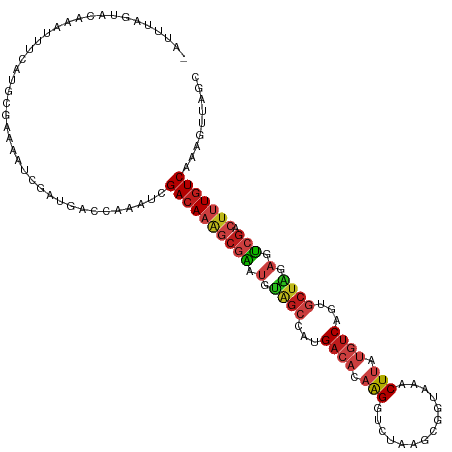
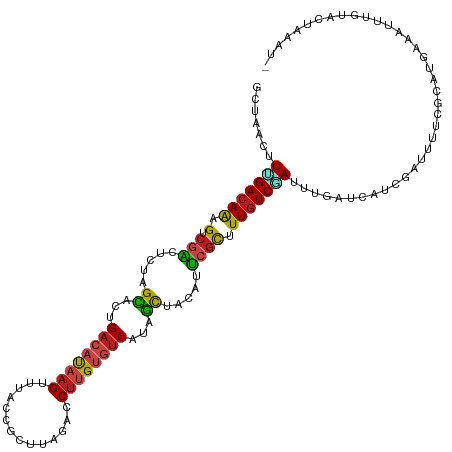
>dm3.chr4 1268748 119 - 1351857 GCUAGCUUAGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAUCAAUUUUCGCAUGAAACUUGUACUAAAU- .........(((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))).(((((((...(((.((((....)))).))).)))))))- ( -33.00, z-score = -3.25, R) >droSim1.chr4 947137 119 - 949497 GCUAGCUUAGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAUCAAUUUUCGCAUGAAACUUGUACUAAAU- .........(((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))).(((((((...(((.((((....)))).))).)))))))- ( -33.00, z-score = -3.25, R) >droSec1.super_40 286808 119 - 298270 GCUAGCUUAGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAUCAAUUUUCGCAUGAAACUUGUACUAAAU- .........(((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))).(((((((...(((.((((....)))).))).)))))))- ( -33.00, z-score = -3.25, R) >droYak2.chr4 1370813 119 - 1374474 GCCAGCUUUGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAUCGAUUUUUGCAUGAAAUUUGUACUAAAU- (((((..(((((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))))).)))))..........((((.......))))......- ( -33.70, z-score = -3.35, R) >droEre2.scaffold_4793 22122 119 + 55562 GCUAGCUUUGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGAUCAUCAAUUUUAGCAUGAAAUUUGUACUAAAU- (((((..(((((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))))).(((.....)))..)))))..................- ( -32.20, z-score = -3.41, R) >droAna3.scaffold_13089 475457 119 - 978470 GCUAAUUUUGACAGAGUCGACUCUAGCACUGACAUAAGUUUACCGCAUAGACCUUGUGUCAUGGCUACAUUCGCUUUGUCAAUUUGAUCAUCAAUUUUCGCGUGGAAUUUAUACUAGAC- ((.....(((((((((.(((.(.((((..(((((((((((((.....)))).)))))))))..)))).).)))))))))))).(((.....))).....))..................- ( -32.20, z-score = -2.90, R) >droPer1.super_51 489175 116 - 524598 GCUAAUUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCACUUAGCCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGGUCGUCGAUUUUUGCAUGGAAUUUAUACUA---- (((((..(((((((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))))))))))).))))).((.((((((.....)))))).))....---- ( -31.40, z-score = -3.35, R) >droWil1.scaffold_181130 16136723 119 + 16660200 GCCAAUUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCGCUCAGCCCUUGUGUCAUGGCUACAUUCGCCUUGUCGAUUUGAUCGUCGAUUUUUGCGUGAAAUUUGUACUGCAU- ((.((..(((((.((((((((........(((((((((..(........)..))))))))).(((.......)))..))))))))....)))))..)).))..................- ( -29.70, z-score = -1.45, R) >droVir3.scaffold_13052 604576 120 - 2019633 GCCAACUUCGACAAAGUCGACUCUAGCACUGACAUAAGUUUGCCACUUAAUCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGAUCAUCGAUUUUUGCGUGGAAUUUGUACUGUUUU .(((...(((((((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))))))))))).(((.....)))........)))............... ( -33.70, z-score = -3.49, R) >droMoj3.scaffold_6498 1841579 119 + 3408170 CCUAACUUCGACAAAGUCGACUCCAGCACUGACAUAAGUUUUCCACUUAAACCUUGUGUCAUGGCUACAUUCGCUUUGUCGAUUUGUUCAUCGAUUCUUGCAUGGAAUUUGUACUAAAA- ...(((.(((((((((.(((....(((..(((((((((.(((......))).)))))))))..)))....))))))))))))...))).((.((((((.....)))))).)).......- ( -33.50, z-score = -4.46, R) >droGri2.scaffold_14822 824072 119 + 1097346 GCCAAUUUUGACAAAGUCGACUCCAGCACUGACAUAAGUUUGCCACUGAGGCCUUGUGUCAUCGCCACAUUCGCUUUGUCAAUUUGAUCAUCAAUUUUCGCAUGAAAUUUGUACUACAA- ..(((..(((((((((.(((.....((..(((((((((...(((.....))))))))))))..)).....)))))))))))).))).....(((.((((....)))).)))........- ( -33.60, z-score = -3.57, R) >apiMel3.Group2 14377538 119 + 14465785 GACAAUUUGGAUAAAGUCGUCUCGAGUACGGUGACCAGUUUGUUGAUCAUUCCUUGGGUCAUGGCAGCCGAAGUCUUCUCUAUCAAAUCGUCGAUUUUCGCGUGAUAUUGGUGCUGGAA- (((.((((.....)))).)))((.((((((((..(((......(((((........))))))))..)))........(..(((((...((.((.....)))))))))..)))))).)).- ( -26.90, z-score = 1.28, R) >triCas2.ChLG5 10946359 114 + 18847211 GAUAGCUUCGACAGUGUGGCUUCCAGGACCGACAUCAGCUUGCUGAUCAUCCCCUGGUUCAUGUUCGCCAGGGUUUUGUCGAUUUGGUCGUCGAUUUUCGUGUGGUAUUGGUGA------ ((((.(..(((.(((.((((..(((((..(((((((((....)))).....(((((((........)))))))...))))).)))))..))))))).)))...).)))).....------ ( -35.70, z-score = -0.61, R) >consensus GCUAACUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGAUCAUCGAUUUUCGCAUGAAAUUUGUACUAAAU_ .......(((((((.(.(((.....((...((((((((..............))))))))...)).....)))).)))))))...................................... (-15.33 = -15.31 + -0.02)
| Location | 1,268,787 – 1,268,907 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Shannon entropy | 0.41251 |
| G+C content | 0.45769 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
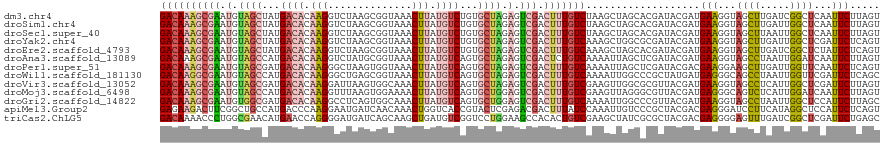
>dm3.chr4 1268787 120 + 1351857 GACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGCACGAUACGAUGAAGGUAGCUUGAUCGGCUCAAUUCUUAGU ((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...(((((...((..((((.(((....))).))))..)).....))))) ( -33.70, z-score = -0.85, R) >droSim1.chr4 947176 120 + 949497 GACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGCACGAUACGAUGAAGGUAGCUUGAUUGGCUCAAUUCUUAGU ((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))(((((((.(.............).))))))).................. ( -31.62, z-score = -0.34, R) >droSec1.super_40 286847 120 + 298270 GACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGCACGAUACGAUGAAGGUAGCUUAAUUGGCUCAAUUCUUAGU ((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))(((((((.(.............).))))))).................. ( -31.92, z-score = -0.68, R) >droYak2.chr4 1370852 120 + 1374474 GACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCAAAGCUGGCGCGAUACGAUGAAGGUAGCUUGAUUGGCUCGAUUCUCAGU ((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...(((((..(((..((((.(((....))).))))..)))....))))) ( -37.60, z-score = -1.46, R) >droEre2.scaffold_4793 22161 120 - 55562 GACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCAAAGCUAGCACGAUACGAUGAAGGUAGCUUGAUCGGCUCUAUUCUCAGU ((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))...........((..((((.(((....))).))))..)).......... ( -33.40, z-score = -0.66, R) >droAna3.scaffold_13089 475496 120 + 978470 GACAAAGCGAAUGUAGCCAUGACACAAGGUCUAUGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUCUGUCAAAAUUAGCUCGAUACGAUGAAGGUAGCCUAAUUGGAUCAAUUCUUAGU ((((.(((((.(.((((..(((((.(((..((....))....))).)))))..)))).).))).)).))))...........(((.((((..(((...))).)))).))).......... ( -28.00, z-score = -0.26, R) >droPer1.super_51 489211 120 + 524598 GACAAAGCGAAUGUAGCGAUGACACAAGGGCUAAGUGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUAGCUCGAUACGACGAAGGAAGCUUGAUUGGUUCAAUUCUCAGU ((((((((((.(.(((((.(((((.(((.(((....)))...))).))))).))))).).))).)))))))...........((..(((..(((....)))..)))..)).......... ( -35.00, z-score = -2.01, R) >droWil1.scaffold_181130 16136762 120 - 16660200 GACAAGGCGAAUGUAGCCAUGACACAAGGGCUGAGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUGGCCCGCUAUGAUGAGGGCAGCCUAAUUGGUUCGAUUCUCAGC ((((((((((.(.((((..(((((.(((.(((....)))...))).)))))..)))).).))).)))))))..(((((((((.(......).))))((((.....)))))))))...... ( -41.70, z-score = -2.34, R) >droVir3.scaffold_13052 604616 120 + 2019633 GACAAAGCGAAUGUAGCGAUGACACAAGGAUUAAGUGGCAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCGAAGUUGGCGCGUUACGAUGAAGGUAGCCUCAUUGGCUCGAUUCUUAGU ((((((((((.(.(((((.(((((.(((..............))).))))).))))).).))).)))))))...((((........))))(((...((((.....))))...)))..... ( -36.34, z-score = -1.40, R) >droMoj3.scaffold_6498 1841618 120 - 3408170 GACAAAGCGAAUGUAGCCAUGACACAAGGUUUAAGUGGAAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCGAAGUUAGGGCGUUACGAUGAGGGCAGUCUCAUUGGAUCAAUUCUUAGU ((((((((((.(.((((..(((((..((((((.......)))))).)))))..)))).).))).)))))))....((((((.....((((((((....)))))))).......)))))). ( -36.30, z-score = -2.16, R) >droGri2.scaffold_14822 824111 120 - 1097346 GACAAAGCGAAUGUGGCGAUGACACAAGGCCUCAGUGGCAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCAAAAUUGGCCCGUUACGAUGAAGGUAGCCUAAUUGGCUCCAUUCUUAGC ((((((((((.(.(((((.(((((.((((((.....)))...))).))))).))))).).))).)))))))...((((........))))(((((.((((.....)))))).)))..... ( -41.30, z-score = -2.58, R) >apiMel3.Group2 14377577 120 - 14465785 GAGAAGACUUCGGCUGCCAUGACCCAAGGAAUGAUCAACAAACUGGUCACCGUACUCGAGACGACUUUAUCCAAAUUGUCCCGCUACGACGAGGGAUCCUUCAUAGGCUCCAUUCUCAGU ((((((....)((..((((((((((......((((((......)))))).((((..((.(((((.(((....)))))))).)).))))....))).....)))).))).)).)))))... ( -30.10, z-score = -0.34, R) >triCas2.ChLG5 10946393 120 - 18847211 GACAAAACCCUGGCGAACAUGAACCAGGGGAUGAUCAGCAAGCUGAUGUCGGUCCUGGAAGCCACACUGUCGAAGCUAUCGCGCUACGACGAGGGGAGUUUGAUCGGCUCGAUUCUGAGC ((((((.(((((((.........(((((((((.(((((....))))))))..))))))..)))....(((((.(((......))).)))))))))...)))).)).(((((....))))) ( -41.70, z-score = -1.32, R) >consensus GACAAAGCGAAUGUAGCCAUGACACAAGGUCUAAGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAGUUAGCACGAUACGAUGAAGGUAGCUUGAUUGGCUCAAUUCUUAGU ((((((((((.(.((((...((((.(((..............))).))))...)))).).))).)))))))...................(((...((((.....))))...)))..... (-22.48 = -23.08 + 0.60)
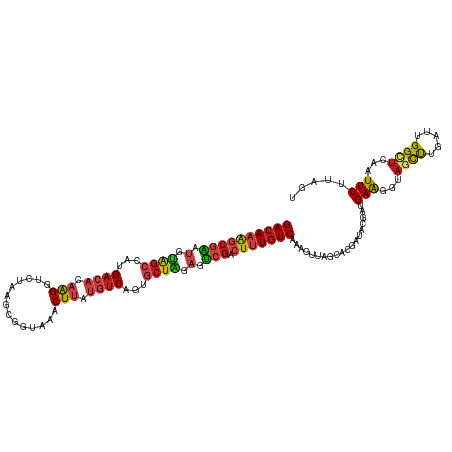
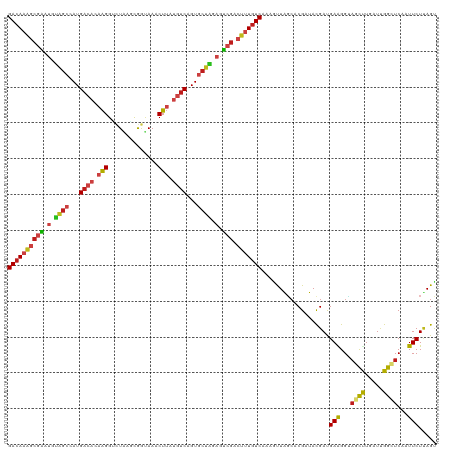
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:58 2011