| Sequence ID | dm3.chr4 |
|---|---|
| Location | 1,021,123 – 1,021,243 |
| Length | 120 |
| Max. P | 0.695733 |

| Location | 1,021,123 – 1,021,243 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.55 |
| Shannon entropy | 0.77060 |
| G+C content | 0.50065 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -8.41 |
| Energy contribution | -9.53 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 1021123 120 + 1351857 AGCAGAGGAAGACACGCCUAGAGGAGUGUUUAUUCCAGAGUUGGCAGAACAGAUGGGACGACGAAAGCGAACCAGGACGGGUGACGCAUAGGUUUAUGCCAUCCGUCACUCUCGCCUAUC ......(((((((((.((....)).)))))..)))).((...((((((.....(((..((.(....)))..))).((((((((..(((((....)))))))))))))..))).)))..)) ( -43.10, z-score = -3.06, R) >droSim1.chr2h_random 2220583 116 + 3178526 UUCACAAGAUGGCGAGACUGAACGAAUGCUUACAUAACGCAUGGCAGCGCAGAUGGUAUAACCCUGAGGAAC----ACAGCCGACUCAUCGGUUCCUCCCAGAAGUAAGAUUCGUUUUCA ...............((..((((((((.(((((....(((......)))..............(((.(((..----..((((((....))))))..))))))..))))))))))))))). ( -29.00, z-score = -0.66, R) >droAna3.scaffold_13278 92127 117 + 130239 GUGAAUGGAAGUCUAUGUUGCGUGAAUGUGUCCUGUCGGACUGGCAAGCCCGAUGGGAUAAUAGCGA-GAAU--GGGCGGGCUACUUACGAGUUCAUUCCAGAUGUCUCGUUUGCUGUCG ..........((((((....(((.....(((((((((((.((....)).)))))))))))...))).-..))--)))).(((..(..(((((..((((...)))).)))))..)..))). ( -39.50, z-score = -1.59, R) >droVir3.scaffold_12945 171572 117 - 206789 GUGAAUGCAAGAGAUUGGUGAAAGAUUGUGUUUUGUCUGACUGGCAAGUCAGAUGGGACGCAAGCGU-GAGU--GGGCGUGUCACUCAGCGGUUUAUACGGGAGGUUACAUUUGCCGGCA .....(((.........(((((...((((((((..(((((((....)))))))..))))))))((.(-((((--((.....)))))))))..))))).(((.((((...)))).)))))) ( -42.10, z-score = -2.83, R) >droGri2.scaffold_15040 45031 117 - 162499 GUGAGUGCAAGAAAUUGCUAAAUGAGUGUGUUCUGUCUGACUGGCAGGUCAGAUGGGAUACUAGCGA-GAAU--GGGAGGGUCACUCAUAGGUUUAUUCGGGAGGUCACAUUUGCUGUUG ((((.(((((....))))...(((((((((((((((((((((....))))))))))))))(((....-...)--))......)))))))..............).))))........... ( -35.60, z-score = -1.64, R) >consensus GUGAAUGGAAGACAAUGCUGAAUGAAUGUGUACUGUCGGACUGGCAGGUCAGAUGGGAUAACAGCGA_GAAU__GGGCGGGUCACUCAUAGGUUUAUUCCAGAGGUCACAUUUGCUGUCA .......................(((((((((((((((((((....))))))))))))))........((((...................)))).............)))))....... ( -8.41 = -9.53 + 1.12)

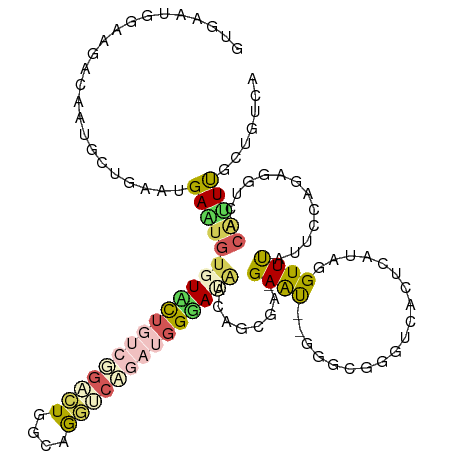

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:52 2011