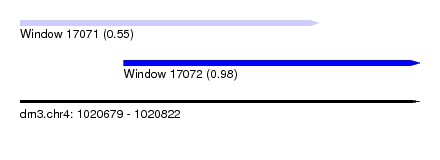
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 1,020,679 – 1,020,822 |
| Length | 143 |
| Max. P | 0.976260 |

| Location | 1,020,679 – 1,020,786 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 59.29 |
| Shannon entropy | 0.66329 |
| G+C content | 0.53362 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -13.44 |
| Energy contribution | -17.75 |
| Covariance contribution | 4.31 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
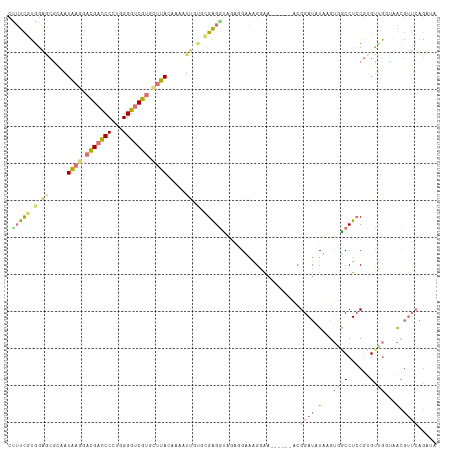
>dm3.chr4 1020679 107 + 1351857 ---UCGUGGGAGAUCAUGCCGACGACCCAUGGGGGCGCGUCUACAAGAUUUGCCGAGGCCGCAGAAAGUGC------ACGGAGAUUGGAUGCCUCCACGUGAAUGGCGAGCUGAUC ---........(((((.((((.(.(..((((((((.((((((....(((((.(((..((.((.....))))------.)))))))))))))))))).))))..).))).))))))) ( -34.60, z-score = 0.37, R) >droAna3.scaffold_13278 90558 112 + 130239 CUUUCGUAGAUCGAAAUAAACACGAUCCCUGGGGUCGUGUUUACAAGAUUCUGCGGGAGAAUGGGAAGGAGGGGUCCAGGGAUAUUUGUGCCCUACGGGUUG--AAGGUUGAGA-- ((..((((((((....((((((((((((...))))))))))))...)).))))))..)).........(.((((..((((....))))..)))).)......--..........-- ( -39.10, z-score = -2.24, R) >droVir3.scaffold_12945 170395 110 - 206789 CUUUCCUGGAGCGCAAUAAGGAUGAUCCCUGGGGUCGUGCUUAUAAAAUAGUUCGAGGUAGAGGCAGGGAA------ACGGAUGUAAGUUGCCUCCGUGCUGGCAACGUUCAGUUA (((((((.((((...(((((.(((((((...))))))).)))))......)))).))).))))(((.(...------.)...)))..((((((........))))))......... ( -37.30, z-score = -1.78, R) >droGri2.scaffold_15040 43474 110 - 162499 CCUUUUUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUGCUUAUAAAGUUGUGAGAGGUAGGCGUAGGGAA------GCGGAUGUAAGUGGCCUCCGUGUCGGUGACGUUCAGUUA .(((((((.(((...(((((.(((((((...))))))).)))))...))).)))))))(..((((...((.------(((((.((.....)).))))).))....))))..).... ( -37.90, z-score = -1.49, R) >consensus CUUUCGUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUGCUUACAAAAUUGUGCGAGGUAGAGGAAAGGAA______ACGGAUAUAAGUGGCCUCCGUGUUGGUAACGUUCAGAUA .((((((((((....(((((.(((((((...))))))).)))))....)))))))))).....................((((((((.((((......)))).))))))))..... (-13.44 = -17.75 + 4.31)

| Location | 1,020,716 – 1,020,822 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 54.83 |
| Shannon entropy | 0.73525 |
| G+C content | 0.48859 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
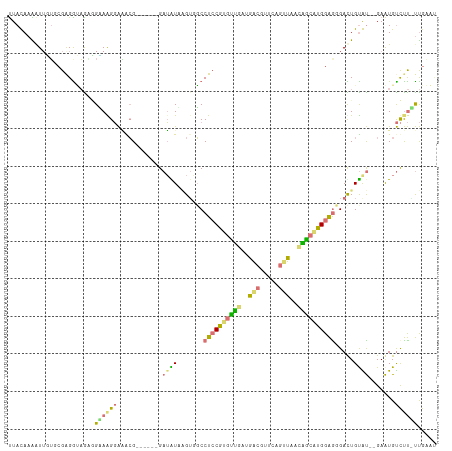
>dm3.chr4 1020716 106 + 1351857 CUACAAGAUUUGCCGAGGCCGCAGAAAGUGCACG------GAGAUUGGAUGCCUCCACGUGAAUGGCGAGCUGAUCACUGAUUGGGGUGACUGUGCACGAGUGCUCCUCCGC-- ...........((.((((..(((....(((((((------(.(((..(.((((...........))))..)..)))(((......)))..))))))))...))).)))).))-- ( -34.00, z-score = -0.19, R) >droAna3.scaffold_13278 90598 112 + 130239 UUACAAGAUUCUGCGGGAGAAUGGGAAGGAGGGGUCCAGGGAUAUUUGUGCCCUACGGGUUGAAGGUUGAGACCUUAUAACCUGGAAGGAUUGUGUU-GAAUGUCUU-UUGUCC .......(((((.....))))).....(((((((..((((....))))..)))).((((((((((((....))))).)))))))(((((((......-....)))))-)).))) ( -30.40, z-score = -0.06, R) >droVir3.scaffold_12945 170435 106 - 206789 UUAUAAAAUAGUUCGAGGUAGAGGCAGGGAAACG------GAUGUAAGUUGCCUCCGUGCUGGCAACGUUCAGUUAACAGCAUGGAGGGAUUGUAU--GAAUGUCUUGUUGAAU ..........((((((....((((((.(....).------...(((...(.(((((((((((..(((.....)))..))))))))))).)...)))--...)))))).)))))) ( -36.30, z-score = -4.25, R) >droGri2.scaffold_15040 43514 106 - 162499 UUAUAAAGUUGUGAGAGGUAGGCGUAGGGAAGCG------GAUGUAAGUGGCCUCCGUGUCGGUGACGUUCAGUUAACGGCAUGGAGUGACUGCAU--GAAUGUCUUAUUGAAU ..........((((((.((..((........)).------.(((((.((.((.(((((((((.((((.....)))).))))))))))).)))))))--..)).))))))..... ( -33.40, z-score = -2.74, R) >consensus UUACAAAAUUGUGCGAGGUAGAGGAAAGGAAACG______GAUAUAAGUGGCCUCCGUGUUGAUGACGUUCAGUUAACAGCAUGGAGGGACUGUAU__GAAUGUCUU_UUGAAU .......................(((((.......................(((((((((((.((((.....)))).)))))))))))....((((....)))))))))..... (-12.74 = -12.05 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:50 2011