
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 1,016,876 – 1,016,994 |
| Length | 118 |
| Max. P | 0.994332 |

| Location | 1,016,876 – 1,016,992 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.09 |
| Shannon entropy | 0.10895 |
| G+C content | 0.33246 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
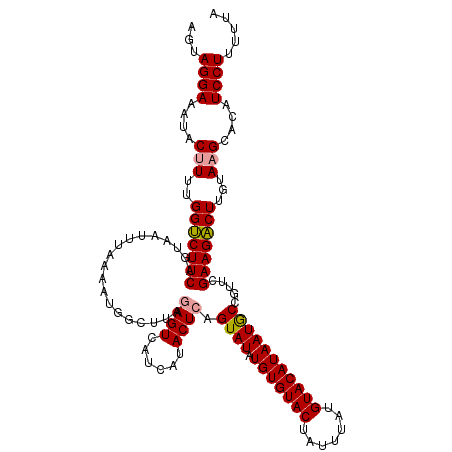
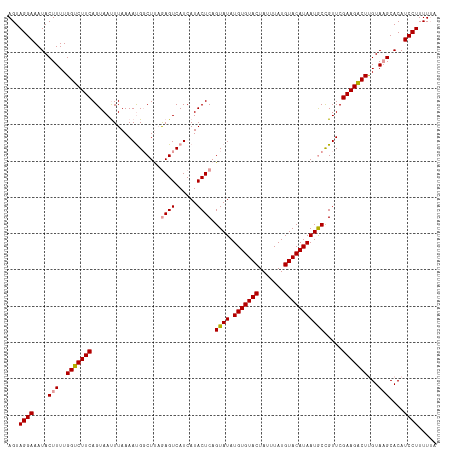
>dm3.chr4 1016876 116 + 1351857 AGUAGGAAAUACUUUUGGCCUUCAGUAAUUUAAAACG-CUCUAAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUACUGUUCGAAGGCUCGUACGCACAUCCUUUUA- ...((((........(((((((((((((((((.....-...))))).....))))((((((.(((((((.......)))))))))))))...)))))).))........))))....- ( -26.79, z-score = -2.56, R) >droSec1.super_40 59901 118 + 298270 AGUAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUA ...((((....(((..((((((((....)....((((((...((((......))))......(((((((.......)))))))..)))))).)))))))...)))....))))..... ( -28.70, z-score = -2.46, R) >droSim1.chr4 786745 118 + 949497 AGUAGGAAAUACUUUUGGUCUUCAGUAACUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUA ...((((....(((..(((((((..........((((((...((((......))))......(((((((.......)))))))..)))))).)))))))...)))....))))..... ( -28.20, z-score = -2.09, R) >consensus AGUAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUA ...((((....(((..(((((((...................((((......)))).((((.(((((((.......))))))))))).....)))))))...)))....))))..... (-25.00 = -25.00 + 0.00)

| Location | 1,016,876 – 1,016,992 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Shannon entropy | 0.10895 |
| G+C content | 0.33246 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -25.52 |
| Energy contribution | -24.97 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
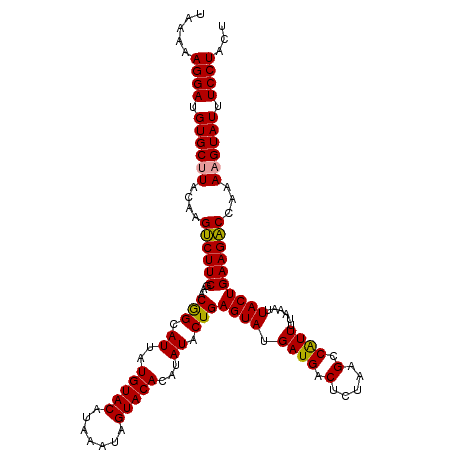
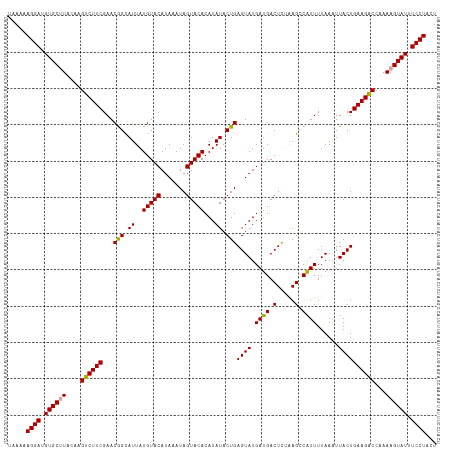
>dm3.chr4 1016876 116 - 1351857 -UAAAAGGAUGUGCGUACGAGCCUUCGAACAGUAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUUAGAG-CGUUUUAAAUUACUGAAGGCCAAAAGUAUUUCCUACU -....((((.((((....(.((((((...((((((..(((((.......)))))...))))))((((....(((......-.)))......)))))))))))....)))).))))... ( -31.50, z-score = -3.67, R) >droSec1.super_40 59901 118 - 298270 UAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUACU .....((((.((((((....(((((((((.(((....(((((.......)))))........((((......))))...))).))).........))))))...)))))).))))... ( -27.70, z-score = -3.22, R) >droSim1.chr4 786745 118 - 949497 UAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAGUUACUGAAGACCAAAAGUAUUUCCUACU .....((((.((((((....((((((.((((((....(((((.......)))))........((((......))))...))).......)))...))))))...)))))).))))... ( -28.11, z-score = -2.85, R) >consensus UAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUACU .....((((.((((((....((((((...(((.((..(((((.......)))))...)).)))((((.((((.(.....).))))......))))))))))...)))))).))))... (-25.52 = -24.97 + -0.55)

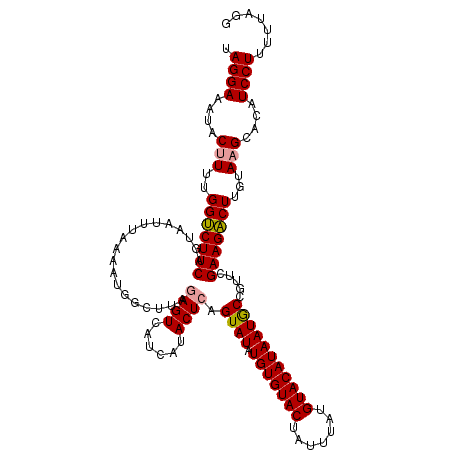
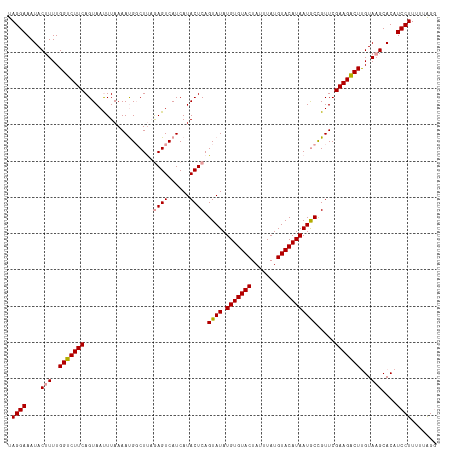
| Location | 1,016,878 – 1,016,994 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Shannon entropy | 0.11673 |
| G+C content | 0.34098 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 1016878 116 + 1351857 UAGGAAAUACUUUUGGCCUUCAGUAAUUUAAAACG-CUCUAAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUACUGUUCGAAGGCUCGUACGCACAUCCUUUUAGG- .((((........(((((((((((((((((.....-...))))).....))))((((((.(((((((.......)))))))))))))...)))))).))........))))......- ( -26.79, z-score = -2.59, R) >droSec1.super_40 59903 118 + 298270 UAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGG .((((....(((..((((((((....)....((((((...((((......))))......(((((((.......)))))))..)))))).)))))))...)))....))))....... ( -28.70, z-score = -2.46, R) >droSim1.chr4 786747 118 + 949497 UAGGAAAUACUUUUGGUCUUCAGUAACUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGG .((((....(((..(((((((..........((((((...((((......))))......(((((((.......)))))))..)))))).)))))))...)))....))))....... ( -28.20, z-score = -2.12, R) >consensus UAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGG .((((....(((..(((((((...................((((......)))).((((.(((((((.......))))))))))).....)))))))...)))....))))....... (-25.00 = -25.00 + 0.00)

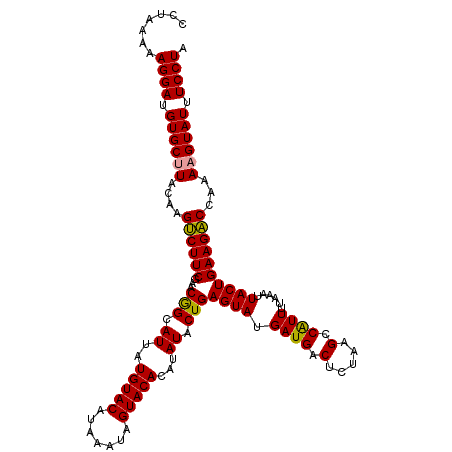
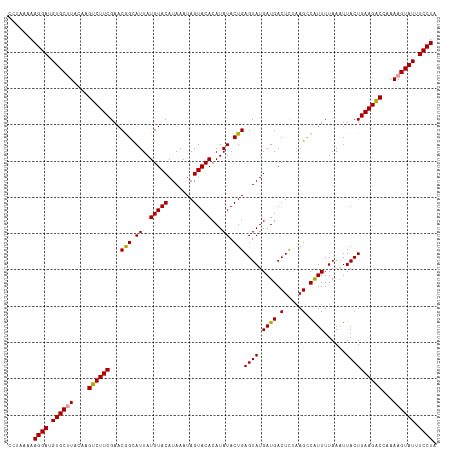
| Location | 1,016,878 – 1,016,994 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Shannon entropy | 0.11673 |
| G+C content | 0.34098 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -25.52 |
| Energy contribution | -24.97 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 1016878 116 - 1351857 -CCUAAAAGGAUGUGCGUACGAGCCUUCGAACAGUAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUUAGAG-CGUUUUAAAUUACUGAAGGCCAAAAGUAUUUCCUA -......((((.((((....(.((((((...((((((..(((((.......)))))...))))))((((....(((......-.)))......)))))))))))....)))).)))). ( -31.50, z-score = -3.69, R) >droSec1.super_40 59903 118 - 298270 CCUAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUA .......((((.((((((....(((((((((.(((....(((((.......)))))........((((......))))...))).))).........))))))...)))))).)))). ( -27.70, z-score = -3.17, R) >droSim1.chr4 786747 118 - 949497 CCUAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAGUUACUGAAGACCAAAAGUAUUUCCUA .......((((.((((((....((((((.((((((....(((((.......)))))........((((......))))...))).......)))...))))))...)))))).)))). ( -28.11, z-score = -2.81, R) >consensus CCUAAAAAGGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUA .......((((.((((((....((((((...(((.((..(((((.......)))))...)).)))((((.((((.(.....).))))......))))))))))...)))))).)))). (-25.52 = -24.97 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:49 2011