
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,406,299 – 9,406,417 |
| Length | 118 |
| Max. P | 0.781614 |

| Location | 9,406,299 – 9,406,417 |
|---|---|
| Length | 118 |
| Sequences | 7 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 68.96 |
| Shannon entropy | 0.58080 |
| G+C content | 0.38354 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -10.79 |
| Energy contribution | -13.03 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
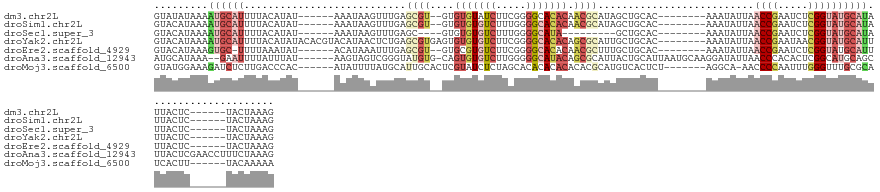
>dm3.chr2L 9406299 118 - 23011544 GUAUAUAAAAUGCAUUUUACAUAU------AAAUAAGUUUGAGCGU--GUGUGUAUCUUCGGGGCACACAACGCAUAGCUGCAC--------AAAUAUUAACCGAAUCUCGGUAUGCAUAUUACUC------UACUAAAG (((..(((.((((((.........------.....((((...((((--((((((.((....)))))))).))))..))))....--------........((((.....)))))))))).)))...------)))..... ( -29.60, z-score = -2.66, R) >droSim1.chr2L 9182598 118 - 22036055 GUACAUAAAAUGCAUUUUACAUAU------AAAUAAGUUUGAGCGU--GUGUGUGUCUUUGGGGCACACAACGCAUAGCUGCAC--------AAAUAUUAACCGAAUCUCGGUAUGCAUAUUACUC------UACUAAAG (((..(((.((((((.........------.....((((...((((--.((((((((.....))))))))))))..))))....--------........((((.....)))))))))).)))...------)))..... ( -30.80, z-score = -2.44, R) >droSec1.super_3 4859924 107 - 7220098 GUACAUAAAAUGCAUUUUACAUAU------AAAUAAGUUUGAGC----GUGUGUGUCUUUGGGGCAUA---------GCUGCAC--------AAAUAUUAACCGAAUCUCGGUAUGCAUAUUACUC------UACUAAAG (((..(((.((((((.........------......(((((.((----(..((((((.....))))))---------..))).)--------))))....((((.....)))))))))).)))...------)))..... ( -23.90, z-score = -1.42, R) >droYak2.chr2L 12077520 126 - 22324452 GUACAUAAAAUGCAUUUUACAUAUACACGUACAUAACUCUGAGCGUGAGUGUGUGUCUUCGGGGCACACAGCGCAUUGCUGCAC--------AAAUAUUAACCGAAUAACGGUAUGCAUUUUACUC------UACUAAAG (((..((((((((((..........(((((.((......)).)))))..((((((((.....))))))))(.(((....))).)--------........((((.....))))))))))))))...------)))..... ( -33.70, z-score = -2.31, R) >droEre2.scaffold_4929 10015398 117 - 26641161 GUACAUAAAGUGC-UUUUAAAUAU------ACAUAAAUUUGAGCGU--GUGCGUGUCUUCGGGGCACACAACGCUUUGCUGCAC--------AAAUAUUAACCGAAUCUCGGUAUGCAUUUUACUC------UACUAAAG (((..((((((((-.....(((((------..........((((((--.((.(((((.....))))).))))))))........--------..))))).((((.....))))..))))))))...------)))..... ( -29.55, z-score = -2.38, R) >droAna3.scaffold_12943 757955 131 - 5039921 AUGCAUAAA--GAAUUUUAUUUAU------AAGUAGUCGGGUAUGUG-CAGUGUGUCUUGGGGGCAUACAGCGCAUUACUGCAUUAAUGCAAGGAUAUUAACCCACACUCGGCAUGCAGCUUACUCGAACCUUUCUAAAG ..(((((((--........)))))------..(((((((((((((((-(.(((((((.....))))))).))))))...((((....)))).((........))..))))))).))).)).................... ( -33.40, z-score = -0.67, R) >droMoj3.scaffold_6500 16293436 120 - 32352404 GUAUGGAAAGAUCUCUUGACCCAC------AUAUUUUAUGCAUUGCACUCGUAUCUCUAGCACACACACACACGCAUGUCACUCU-------AGGCA-AACCCCAAUUUGGGUUUGCGCAUCACUU------UACAAAAA ((((((.(((....)))...))).------.......((((...............((((...(((..(....)..)))....))-------))(((-(((((......)))))))))))).....------)))..... ( -22.90, z-score = -1.04, R) >consensus GUACAUAAAAUGCAUUUUACAUAU______AAAUAAGUUUGAGCGU__GUGUGUGUCUUCGGGGCACACAACGCAUUGCUGCAC________AAAUAUUAACCGAAUCUCGGUAUGCAUAUUACUC______UACUAAAG .........((((((...........................((((....(((((((.....))))))).))))..........................((((.....))))))))))..................... (-10.79 = -13.03 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:14 2011