| Sequence ID | dm3.chr4 |
|---|---|
| Location | 994,197 – 994,272 |
| Length | 75 |
| Max. P | 0.750346 |

| Location | 994,197 – 994,272 |
|---|---|
| Length | 75 |
| Sequences | 11 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Shannon entropy | 0.34531 |
| G+C content | 0.40322 |
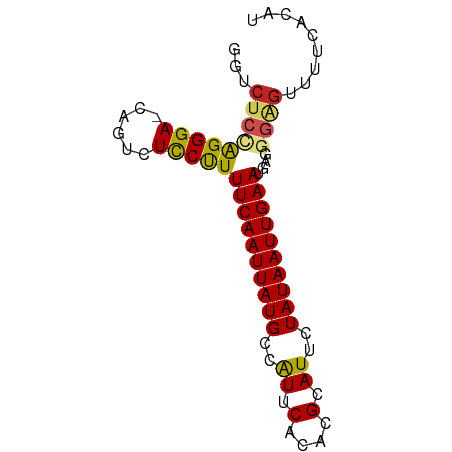
| Mean single sequence MFE | -18.09 |
| Consensus MFE | -10.53 |
| Energy contribution | -10.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750346 |
| Prediction | RNA |
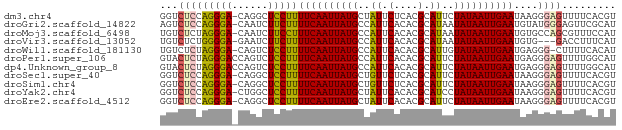
Download alignment: ClustalW | MAF
>dm3.chr4 994197 75 - 1351857 GGUCUCCAGGGA-CAGGCUCCUUUUCAAUUAUGCUAUUCUCACGCAUUCUAUAAUUGAAUAAGGGAGUUUUCACGU .((((....)))-)(((((((((((((((((((..((.(....).))..)))))))))..))))))))))...... ( -20.60, z-score = -3.00, R) >droGri2.scaffold_14822 975421 75 + 1097346 AGUCUCCAGGGA-CAAUCUUCUUUUCAAUUAUGCCAUUCACACGCAUAAUAUAAUUGAAUGUAUGGGAGUUCGCAU .((((....)))-)...(((((.((((((((((..((.(....).))..)))))))))).....)))))....... ( -13.50, z-score = -0.65, R) >droMoj3.scaffold_6498 2044224 75 + 3408170 UGUCUCUAGGGA-CAAUCUUCCUUUCAAUUAUGCCAUUCACACGCAUAAUAUAAUUGAAUGUGCCAGCGUUUCCAU (((((....)))-))............(((((((.........)))))))......((((((....)))))).... ( -12.60, z-score = -0.89, R) >droVir3.scaffold_13052 853993 72 - 2019633 UGUCUCUGGGGA-GAAUCUUCUUUUCAAUUAUGCCAUUCACACGCAUAAUAUAAUUGAAUGUG---GACCUUUCAU .((((.((..((-((....))))..))(((((((.........)))))))............)---)))....... ( -14.10, z-score = -0.94, R) >droWil1.scaffold_181130 1255531 74 - 16660200 UGUCUCUAGGGA-CAGUCUCCUUUUCAAUUAUGCCAUUCACACGCAUUGUAUAAUUGAAUGAGGG-CUUUUCACAU (((((....)))-))...((((((((((((((((.((.(....).)).))))))))))).)))))-.......... ( -19.30, z-score = -2.73, R) >droPer1.super_106 30693 76 - 146283 GUACUCUAGGGACCAGUCUCCUUUUCAAUUAUGCCAUUCACACGCAUUCUAUAAUUGAAUGAGGGAGUUUUGGCAU ..........(.((((.((((((((((((((((..((.(....).))..)))))))))..)))))))..))))).. ( -17.50, z-score = -1.27, R) >dp4.Unknown_group_8 194217 76 + 194412 GUACUCUAGGGACCAGUCUCCUUUUCAAUUAUGCCAUUCACACGCAUUCUAUAAUUGAAUGAGGGAGUUUUGGCAU ..........(.((((.((((((((((((((((..((.(....).))..)))))))))..)))))))..))))).. ( -17.50, z-score = -1.27, R) >droSec1.super_40 35801 75 - 298270 GGUCUCCAGGGA-CAGGCUCCUUUUCAAUUAUGCUGUUCUCACGCAUUCUAUAAUUGAAUAAGGGAGUUUUCACGU .((((....)))-)(((((((((((((((((((.(((......)))...)))))))))..))))))))))...... ( -21.60, z-score = -2.86, R) >droSim1.chr4 771482 75 - 949497 GGUCUCCAGGGA-CAGGCUCCUUUUCAAUUAUGCUGUUCUCACGCAUUCUAUAAUUGAAUAAGGGAGUUUUCACGU .((((....)))-)(((((((((((((((((((.(((......)))...)))))))))..))))))))))...... ( -21.60, z-score = -2.86, R) >droYak2.chr4 1059937 75 - 1374474 GGUCUCCAGGGA-CUGGCUCCUUUUCAAUUAUGCUAUUCACACGCAUCCUAUAAUUGAAUAAGGGAGUUUUCACGU (((((....)))-))((((((((((((((((((..((.(....).))..)))))))))..)))))))))....... ( -20.00, z-score = -2.36, R) >droEre2.scaffold_4512 1002113 75 + 1286254 GGUCUCCAGGGA-CAGGCUCCUUUUCAAUUAUGCUAUUCACACGCAUUCUAUAAUUGAAUAAGGGAGUUUUCACGU .((((....)))-)(((((((((((((((((((..((.(....).))..)))))))))..))))))))))...... ( -20.70, z-score = -3.29, R) >consensus GGUCUCCAGGGA_CAGUCUCCUUUUCAAUUAUGCCAUUCACACGCAUUCUAUAAUUGAAUGAGGGAGUUUUCACAU ...(((((((((......)))))((((((((((..((.(....).))..))))))))))....))))......... (-10.53 = -10.55 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:45 2011