
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 954,350 – 954,476 |
| Length | 126 |
| Max. P | 0.999787 |

| Location | 954,350 – 954,476 |
|---|---|
| Length | 126 |
| Sequences | 5 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 52.15 |
| Shannon entropy | 0.78965 |
| G+C content | 0.43871 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
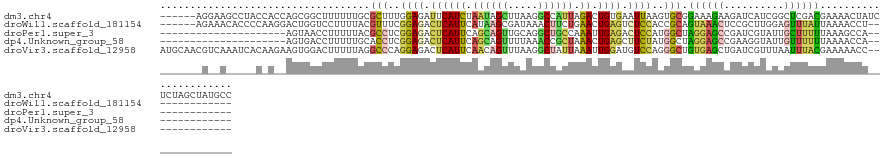
>dm3.chr4 954350 126 + 1351857 ------AGGAAGCCUACCACCAGCGGCUUUUUUGCGCUUUGGAGAUUCAUCUAAUAGCUUAAGGCCAUUAGACUGUGAAUUAAGUGCGGAAAGAAGAUCAUCGGCUCGACGAAAACUAUCUCUAGCUAUGCC ------..((((((..........))))))(((((((((....((((((((((((.((.....)).))))))...)))))))))))))))........(((.((((.((.((......)))).))))))).. ( -30.60, z-score = 0.38, R) >droWil1.scaffold_181154 1898783 112 - 4610121 ------AGAAACACCCCAAGGACUGGUCCUUUUACGUUUCGGAGACUCAUUCAUAAGCGAUAAACUUCUGAACUGAGUCUCCACCGCAGUAAACUCCGCUUGGAGUUUAUUAAAACCU-------------- ------.(((((.....(((((....)))))....)))))(((((((((((((.(((.......))).)))).))))))))).....((((((((((....)))))))))).......-------------- ( -40.60, z-score = -5.99, R) >droPer1.super_3 489144 97 + 7375914 ---------------------AGUAACCUUUUUACGCCUCGGAGACUCAUUCAGCAGUUGCAGGCUGCCAAAUUGAGACUCCAUGGCUAGGAGCCGAUCGUAUUGCUUUUUAAAGCCA-------------- ---------------------.((((.....)))).....((((.((((....(((((.....))))).....)))).)))).(((((((((((..........))))))...)))))-------------- ( -27.40, z-score = -1.14, R) >dp4.Unknown_group_58 28531 97 - 69560 ---------------------AGUGACCUUUUUGCACCUCGGAGACUCAUUCAGCAGUUUUAAACCGCUAAACUGAGCUUCUAUGGCUAGGAGCCGAAGGUAUUGUUUUUUAAAACCA-------------- ---------------------....((((((.........((((.((((((.(((.((.....)).))).)).))))))))...(((.....))))))))).................-------------- ( -21.30, z-score = -0.03, R) >droVir3.scaffold_12958 1584457 118 - 3547706 AUGCAACGUCAAAUCACAAGAAGUGGACUUUUUAGGCCCAGGAGACUCAUUCAACAGUUUAAGGCUAUUAAAUUGGAUGUCCAGGGCUGUGAGCUGAUCGUUUAAUUUACGAAAAACC-------------- ....(((((((..((((((((((....)))))).(((((.(((....((((((...((((((.....))))))))))))))).)))))))))..))).))))................-------------- ( -31.20, z-score = -1.77, R) >consensus _______G__A_____C_A__AGUGGCCUUUUUACGCCUCGGAGACUCAUUCAACAGUUUAAGGCUACUAAACUGAGACUCCAGGGCUGGGAGCCGAUCGUAUAGUUUAUUAAAACCA______________ ...................................(((..((((.((((((.((((((.....)))))).)).)))).))))..))).((((((..........))))))...................... (-10.68 = -11.20 + 0.52)

| Location | 954,350 – 954,476 |
|---|---|
| Length | 126 |
| Sequences | 5 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 52.15 |
| Shannon entropy | 0.78965 |
| G+C content | 0.43871 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
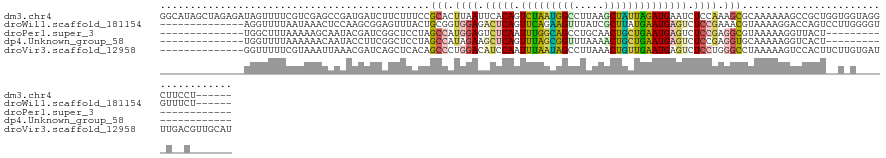
>dm3.chr4 954350 126 - 1351857 GGCAUAGCUAGAGAUAGUUUUCGUCGAGCCGAUGAUCUUCUUUCCGCACUUAAUUCACAGUCUAAUGGCCUUAAGCUAUUAGAUGAAUCUCCAAAGCGCAAAAAAGCCGCUGGUGGUAGGCUUCCU------ ((...(((((((((.((...((((((...)))))).)))))))((((.....(((((...(((((((((.....))))))))))))))......(((((......).)))).))))..)))).)).------ ( -34.00, z-score = -0.64, R) >droWil1.scaffold_181154 1898783 112 + 4610121 --------------AGGUUUUAAUAAACUCCAAGCGGAGUUUACUGCGGUGGAGACUCAGUUCAGAAGUUUAUCGCUUAUGAAUGAGUCUCCGAAACGUAAAAGGACCAGUCCUUGGGGUGUUUCU------ --------------.........((((((((....)))))))).......(((((((((.((((.((((.....)))).)))))))))))))(((((((..(((((....)))))...))))))).------ ( -42.10, z-score = -4.25, R) >droPer1.super_3 489144 97 - 7375914 --------------UGGCUUUAAAAAGCAAUACGAUCGGCUCCUAGCCAUGGAGUCUCAAUUUGGCAGCCUGCAACUGCUGAAUGAGUCUCCGAGGCGUAAAAAGGUUACU--------------------- --------------..((((....))))..((((.(((((.....)))..((((.((((.((..((((.......))))..)))))).))))))..))))...........--------------------- ( -34.00, z-score = -2.80, R) >dp4.Unknown_group_58 28531 97 + 69560 --------------UGGUUUUAAAAAACAAUACCUUCGGCUCCUAGCCAUAGAAGCUCAGUUUAGCGGUUUAAAACUGCUGAAUGAGUCUCCGAGGUGCAAAAAGGUCACU--------------------- --------------..((((....))))..((((((.(((.....)))...((..((((.(((((((((.....)))))))))))))..)).)))))).............--------------------- ( -28.30, z-score = -2.35, R) >droVir3.scaffold_12958 1584457 118 + 3547706 --------------GGUUUUUCGUAAAUUAAACGAUCAGCUCACAGCCCUGGACAUCCAAUUUAAUAGCCUUAAACUGUUGAAUGAGUCUCCUGGGCCUAAAAAGUCCACUUCUUGUGAUUUGACGUUGCAU --------------................((((.((((.(((((((((.(((.(..((.((((((((.......))))))))))..).))).)))).....(((....)))..))))).)))))))).... ( -30.10, z-score = -2.25, R) >consensus ______________UGGUUUUAAUAAACAAUACGAUCAGCUCACAGCCAUGGAGACUCAGUUUAGCAGCCUAAAACUGCUGAAUGAGUCUCCGAGGCGUAAAAAGGCCACU__U_G_____U__C_______ .............................................(((.((((.(((((.(((((((((.....)))))))))))))).)))).)))................................... (-16.92 = -17.32 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:44 2011