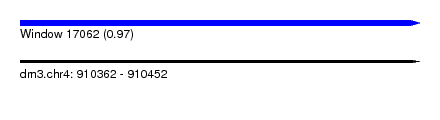
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 910,362 – 910,452 |
| Length | 90 |
| Max. P | 0.966606 |

| Location | 910,362 – 910,452 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 60.60 |
| Shannon entropy | 0.82512 |
| G+C content | 0.38521 |
| Mean single sequence MFE | -15.13 |
| Consensus MFE | -5.46 |
| Energy contribution | -5.12 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
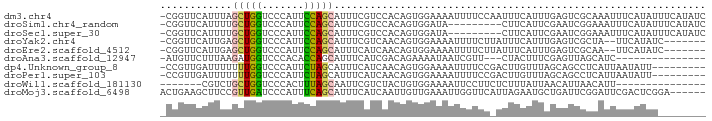
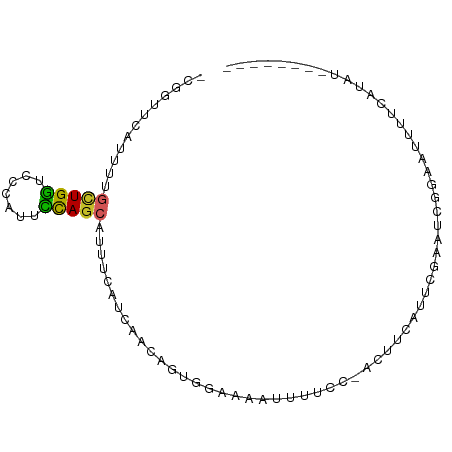
>dm3.chr4 910362 90 + 1351857 -CGGUUCAUUUAGCUGGUCCCAUUCCAGCAUUUCGUCCACAGUGGAAAAUUUUCCAAUUUCAUUUGAGUCGCAAAUUUCAUAUUUCAUAUC -((..(((....(((((.......))))).............(((((....)))))........)))..)).................... ( -17.40, z-score = -3.00, R) >droSim1.chr4_random 104523 81 + 134295 -CGGUUCAUUUUGCUGGUCCCAUUCCAGCAUUUCGUCCACAGUGGAUA---------CUUCAUUCGAAUCGGAAAUUUCAUAUUUCAUAUC -((((((....((((((.......))))))....((((.....)))).---------........))))))(((((.....)))))..... ( -22.20, z-score = -4.16, R) >droSec1.super_30 620163 81 + 664411 -CGGUUCAUUUUGCUGGUCCCAUUCCAGCAUUUCGUCCACAGUGGAUA---------CUUCAUUCGAAUCGGAAAUUUCAUAUUUCAUAUC -((((((....((((((.......))))))....((((.....)))).---------........))))))(((((.....)))))..... ( -22.20, z-score = -4.16, R) >droYak2.chr4 977678 81 + 1374474 -CGGUUCAUUGAGCUGGUCCCAUUCCAGCAUUUCGUCAACAGUGGAAAAUUUUCUUAUUUCAUUUGAGUCGCUA--UUCAUAUC------- -((..(((.((((((((.......))))).(((((.(....))))))............)))..)))..))...--........------- ( -15.80, z-score = -1.56, R) >droEre2.scaffold_4512 925972 81 - 1286254 -CGGUUCAUUGAGCUGGUCCCAUUCCAGCAUUUCAUCAACAGUGGAAAAUUUUCUUAUUUCAUUUGAGUCGCAA--UUCAUAUC------- -((..(((.((((((((.......))))).((((((.....))))))............)))..)))..))...--........------- ( -17.90, z-score = -2.54, R) >droAna3.scaffold_12947 737678 72 + 1574637 -AUGUUCUUUAAGAUGGUCCCACACCAGCAUUUCAUCGACAGAAAAUAAUCGUU---CUACUUUCGAGUUAGCAUC--------------- -.............((((.....))))((......((((.((((........))---))....))))....))...--------------- ( -7.50, z-score = 0.58, R) >dp4.Unknown_group_8 125779 81 - 194412 -CCGUUGAUUUUUUUGGUCCCAUUCUAGCAUUUCAUCAACAGUGGAAAAUUUUCCGACUUGUUUAGCAGCCUCAUUAAUAUU--------- -..((((((....((((.......))))......))))))(((((((....)))).))).......................--------- ( -8.90, z-score = 0.62, R) >droPer1.super_103 103361 81 - 157294 -CCGUUGAUUUUUUUGGUCCCAUUCUAGCAUUUCAUCAACAGUGGAAAAUUUUCCGACUUGUUUAGCAGCCUCAUUAAUAUU--------- -..((((((....((((.......))))......))))))(((((((....)))).))).......................--------- ( -8.90, z-score = 0.62, R) >droWil1.scaffold_181130 15901793 69 - 16660200 -------CGUCUGCUGGUCCCACUUUAGCAAUUCGUCUACUGUGGAAAAUUCCUUCUCUUUAUUAACAUUAACAUU--------------- -------............((((..((((.....).)))..))))...............................--------------- ( -7.00, z-score = -0.18, R) >droMoj3.scaffold_6498 787542 85 + 3408170 ACUGAAGCUUCCGUUGAUCCCAUUUCAGCAUUUCAUCAAUUGUUGAAAUUGGUUCAUUAGAAUGCUGAUUCGGAUUCGACUCGGA------ .........((((((((..((...(((((((((.(((((((.....)))))))......)))))))))...))..)))))..)))------ ( -23.50, z-score = -1.78, R) >consensus _CGGUUCAUUUUGCUGGUCCCAUUCCAGCAUUUCAUCAACAGUGGAAAAUUUUCC_ACUUCAUUCGAAUCGGAAUUUUCAUAU________ ............(((((.......))))).............................................................. ( -5.46 = -5.12 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:42 2011