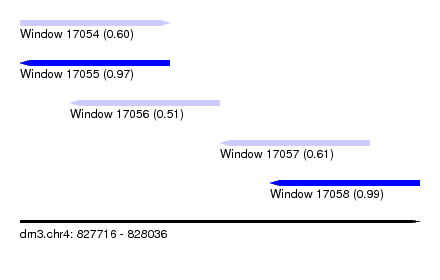
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 827,716 – 828,036 |
| Length | 320 |
| Max. P | 0.994512 |

| Location | 827,716 – 827,836 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Shannon entropy | 0.31209 |
| G+C content | 0.51809 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.63 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 827716 120 + 1351857 CAACACUGUCCCUCUAACGCCACCGACCAAAUAUUUUUUUGUUCGGUGUUCGUUGCUUAGUGCUCAGUUCUGCGGGCAGCGUUUUGUAUACAUACGCUGUGAAAUCAGGCAGAGGGUGUC ....((...((((((((((.((((((.((((......)))).))))))..))))((((..(((........))).(((((((..((....)).)))))))......)))))))))).)). ( -40.90, z-score = -3.00, R) >droEre2.scaffold_4512 845416 94 - 1286254 CCACACCGUCCCUCCACCGCCACCGACCA------------------GUUUGUC------UGUUUUGUUCUGCGGGCAGCGUUCGGUAUACAUACGCUGUGAAAUCGGGCAGUGUAUC-- ..((((.((((.....((((.((.((.((------------------(.....)------)).)).))...))))(((((((..(.....)..)))))))......)))).))))...-- ( -29.90, z-score = -1.33, R) >droSec1.super_30 532993 119 + 664411 CAACACUGUCCCUCCACCGCCACCGACCACAUAUUUUUU-GUUCGGUGUUCGUUGCUUAGUGCUCAGCUCUGCGGGCAGCGUUUGGUAUACAUACGCUGUUAAAUCAGGCAGAGGGUAUC .........(((((..((((((((((..(((.......)-))))))))......(((........)))...))))(((((((.((.....)).)))))))...........))))).... ( -37.60, z-score = -1.84, R) >droSim1.chr4 638864 119 + 949497 CAACACUGUCCCUCCACCGCCACCGACCACAUAUUUUUU-GUUCGGUGUUCGUUGCUUAGUGCUCAGCUCUGCGGGCAGCGUUUGGUAUACAUACGCUGUUAAAUCAGGCAGAGGGUAUC .........(((((..((((((((((..(((.......)-))))))))......(((........)))...))))(((((((.((.....)).)))))))...........))))).... ( -37.60, z-score = -1.84, R) >consensus CAACACUGUCCCUCCACCGCCACCGACCACAUAUUUUUU_GUUCGGUGUUCGUUGCUUAGUGCUCAGCUCUGCGGGCAGCGUUUGGUAUACAUACGCUGUGAAAUCAGGCAGAGGGUAUC .........(((((....(((.((((.(((...............))).))..........((........))))(((((((.((.....)).))))))).......))).))))).... (-21.20 = -22.63 + 1.44)

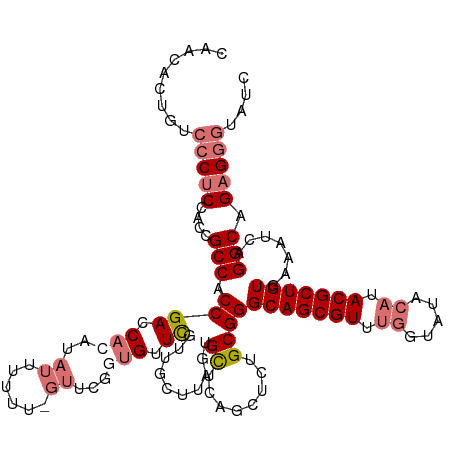
| Location | 827,716 – 827,836 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Shannon entropy | 0.31209 |
| G+C content | 0.51809 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 827716 120 - 1351857 GACACCCUCUGCCUGAUUUCACAGCGUAUGUAUACAAAACGCUGCCCGCAGAACUGAGCACUAAGCAACGAACACCGAACAAAAAAAUAUUUGGUCGGUGGCGUUAGAGGGACAGUGUUG (((((((((((..........((((((.((....))..))))))..(((........((.....))......((((((.((((......)))).))))))))).)))))))....)))). ( -36.00, z-score = -2.32, R) >droEre2.scaffold_4512 845416 94 + 1286254 --GAUACACUGCCCGAUUUCACAGCGUAUGUAUACCGAACGCUGCCCGCAGAACAAAACA------GACAAAC------------------UGGUCGGUGGCGGUGGAGGGACGGUGUGG --..((((((((((.......((((((.((.....)).))))))(((((...((....((------(.....)------------------))....)).)))).)..))).))))))). ( -34.00, z-score = -1.79, R) >droSec1.super_30 532993 119 - 664411 GAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAGCUGAGCACUAAGCAACGAACACCGAAC-AAAAAAUAUGUGGUCGGUGGCGGUGGAGGGACAGUGUUG ....(((((..((........((((((.((.....)).))))))...((...(((........)))......((((((((-(.......)))..))))))))))..)))))......... ( -41.00, z-score = -2.65, R) >droSim1.chr4 638864 119 - 949497 GAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAGCUGAGCACUAAGCAACGAACACCGAAC-AAAAAAUAUGUGGUCGGUGGCGGUGGAGGGACAGUGUUG ....(((((..((........((((((.((.....)).))))))...((...(((........)))......((((((((-(.......)))..))))))))))..)))))......... ( -41.00, z-score = -2.65, R) >consensus GAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAACUGAGCACUAAGCAACGAACACCGAAC_AAAAAAUAUGUGGUCGGUGGCGGUGGAGGGACAGUGUUG ....(((((((((........((((((.((.....)).))))))...((...(((((.(((.............................))).))))).)))))))))))......... (-27.90 = -28.90 + 1.00)

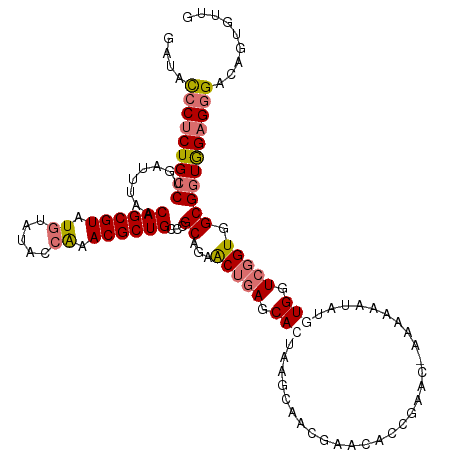
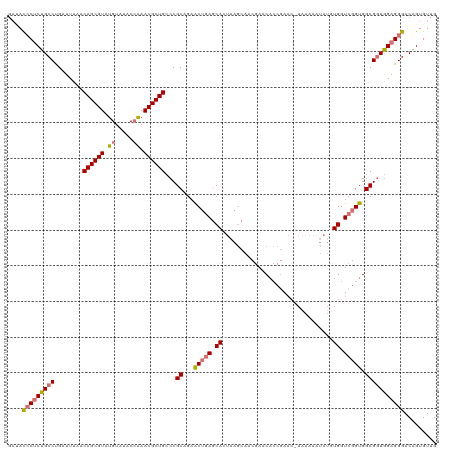
| Location | 827,756 – 827,876 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.30793 |
| G+C content | 0.52626 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -22.08 |
| Energy contribution | -23.32 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 827756 120 - 1351857 GAAGCUAGAGCCCCGAGGGUCAGAGCGGAACAGUUGUUUGGACACCCUCUGCCUGAUUUCACAGCGUAUGUAUACAAAACGCUGCCCGCAGAACUGAGCACUAAGCAACGAACACCGAAC ...(((...((((...))))...)))((....(((((((((......(((((.((....))((((((.((....))..))))))...)))))........))))))))).....)).... ( -32.54, z-score = -1.03, R) >droYak2.chr4 891621 99 - 1374474 GAUGUUAAAGCACCGAGGGUCACAGCAGAACAGUUA--CGGAUACCC--UGCCCGAUUUCACAGCGUAUGUAUACCGAACGCUGUCCGGAGAACAAAACAGAC----------------- ..((((....(.(((.((((...(((......))).--.((....))--.))))......(((((((.((.....)).))))))).))).).....))))...----------------- ( -21.40, z-score = -0.28, R) >droSec1.super_30 533032 120 - 664411 GAAGCUAAAGGCCCGAGGGUCAGAGCAGAACAGUUGCUCGGAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAGCUGAGCACUAAGCAACGAACACCGAAC ...((...((((..((((((..((((((.....))))))....)))))).)))).......((((((.((.....)).))))))...))...(((........))).............. ( -37.40, z-score = -2.37, R) >droSim1.chr4 638903 120 - 949497 GAAGCUAAAGGCCCGAGGGUCAGAGCAGAACAGUUGCUCGGAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAGCUGAGCACUAAGCAACGAACACCGAAC ...((...((((..((((((..((((((.....))))))....)))))).)))).......((((((.((.....)).))))))...))...(((........))).............. ( -37.40, z-score = -2.37, R) >consensus GAAGCUAAAGCCCCGAGGGUCAGAGCAGAACAGUUGCUCGGAUACCCUCUGCCUGAUUUAACAGCGUAUGUAUACCAAACGCUGCCCGCAGAACUGAGCACUAAGCAACGAACACCGAAC ...(((........((((((..((((((.....))))))....))))))...(((......((((((.((.....)).))))))....))).....)))..................... (-22.08 = -23.32 + 1.25)

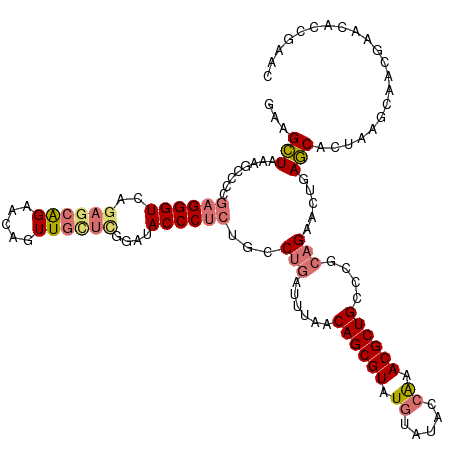
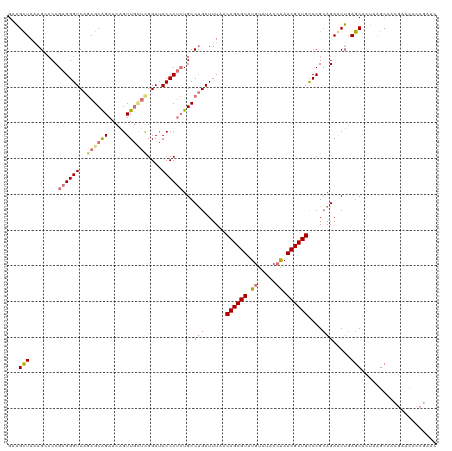
| Location | 827,876 – 827,996 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Shannon entropy | 0.09171 |
| G+C content | 0.56500 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -39.98 |
| Energy contribution | -40.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 827876 120 - 1351857 UGUAGUAGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGAAGACGAGGGCUACCCUAAGUGCGCAAAGAACGGCAAGUUGAUUCG (((.((.((.....)).((((((((..(((((.((((....))).).))))).((((((((....))))))))...))))((((...))))....)))).....)).))).......... ( -38.30, z-score = -0.24, R) >droEre2.scaffold_4512 845535 120 + 1286254 UGUAGUCGGAUAUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAUGGCGUCUGCGACGACGAGAGCUACUCUAAGUGCGCACAGAACGACAAGUGGAUUCG .((((((((.....))..(((((((..(((((.((((....))).).))))).((((((((....))))))))))))))).)).))))((((..((((.......)).))..)))).... ( -44.30, z-score = -1.51, R) >droYak2.chr4 891720 120 - 1374474 UGUAGUCGGAUGUUCCAGCGUCGUCAGUUACCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACGAGGGCUACCCUAAGAGCGCAAGGAAUGACAAGUAGAUUCG ....(((....(((((..(((((((......((((((....)))..)))....((((((((....))))))))))))))).(.(((........))).)..))))))))........... ( -41.70, z-score = -1.20, R) >droSec1.super_30 533152 120 - 664411 UGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGUCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACGAGGGCUACCCUAAGUGCGCAAAGAACGGCAAGUUGAUUCG .((((((((.....))..(((((((..(((((.((((....))).).))))).((((((((....)))))))))))))))..))))))..............((((((....))).))). ( -43.60, z-score = -1.49, R) >droSim1.chr4 639023 120 - 949497 UGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACGAGGGCUACCCUAAGUGCGCAAAGAACGGCAAGUUGAUUCG .((((((((.....))..(((((((..(((((.((((....))).).))))).((((((((....)))))))))))))))..))))))..............((((((....))).))). ( -43.60, z-score = -1.16, R) >consensus UGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACGAGGGCUACCCUAAGUGCGCAAAGAACGGCAAGUUGAUUCG ....(((((.....)).((((((((..(((((.((((....))).).))))).((((((((....)))))))))))))).((((...))))......)).......)))........... (-39.98 = -40.02 + 0.04)

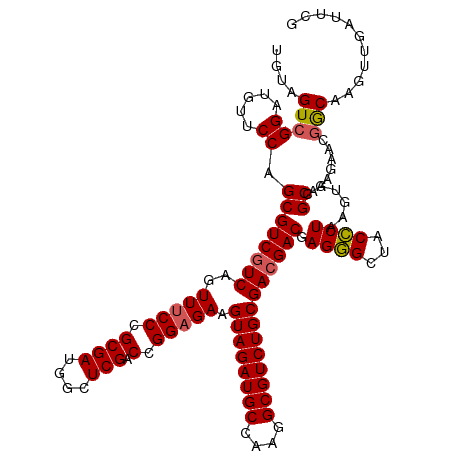
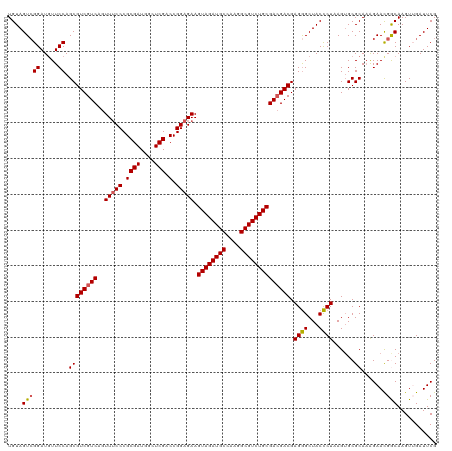
| Location | 827,916 – 828,036 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.40 |
| Shannon entropy | 0.26634 |
| G+C content | 0.56313 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -33.85 |
| Energy contribution | -34.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 827916 120 - 1351857 GUCCAUUGUAAAACUCUGACAUCACAUCAGCACCCUCAGCUGUAGUAGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGAAGACG (((((((((......(((((...((..((((.......))))..))..(((((....))))))))))......)))))).(((.....)))..((((((((....))))))))...))). ( -39.80, z-score = -1.89, R) >droEre2.scaffold_4512 845575 92 + 1286254 ----------------------------UUCAGCCUCAGCUGUAGUCGGAUAUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAUGGCGUCUGCGACGACG ----------------------------..((((....))))..((.((.....)).))((((((..(((((.((((....))).).))))).((((((((....)))))))))))))). ( -38.00, z-score = -2.64, R) >droSec1.super_30 533192 113 - 664411 -------GUAAUGCUCUGACAUCACUUCAGCAUCCUCAGCUGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGUCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACG -------.....(((..(((((((((.((((.......)))).)))..))))))..)))((((((..(((((.((((....))).).))))).((((((((....)))))))))))))). ( -48.10, z-score = -4.14, R) >droSim1.chr4 639063 113 - 949497 -------GUAAUGCUCUGACAUCACAUCAGCAUCCUCAGCUGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACG -------.....(((..((((((....((((.......))))......))))))..)))((((((..(((((.((((....))).).))))).((((((((....)))))))))))))). ( -46.50, z-score = -3.25, R) >consensus _______GUAAUGCUCUGACAUCACAUCAGCAUCCUCAGCUGUAGUCGGAUGUUCCAGCGUCGUCAGUUUCCCGCGAUGGCUCGACCGGAGAAGUAGAUGCCAAGGCGUCUGCGACGACG ...........................((((.......))))..((.((.....)).))((((((..(((((.((((....))).).))))).((((((((....)))))))))))))). (-33.85 = -34.85 + 1.00)

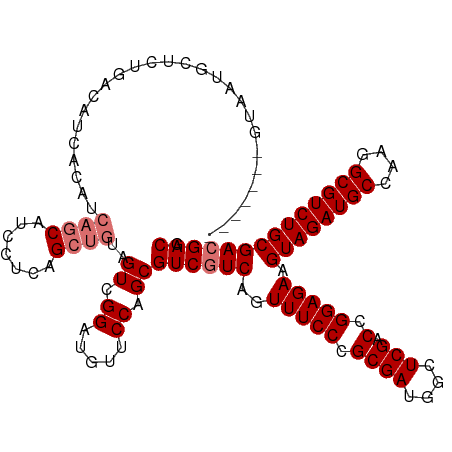
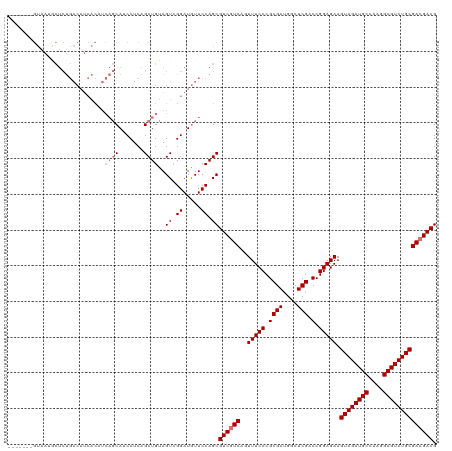
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:39 2011