
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 789,272 – 789,389 |
| Length | 117 |
| Max. P | 0.746049 |

| Location | 789,272 – 789,389 |
|---|---|
| Length | 117 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.59453 |
| G+C content | 0.37496 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -15.08 |
| Energy contribution | -13.23 |
| Covariance contribution | -1.86 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 789272 117 - 1351857 ---AGUAUAUUUUGGUUGUUGCUUACAAUUCCAGUAUUAGGAUCAUUUCAUCGUCGUCUUCGAAAGCAUCGUGUAAGUUGAUAAGCUUCUGAUGAUGCAGUUGAUUCAUUAUAUCUAUUU ---.(((((..((((..((((....)))).)))).....((((((........(((....)))..(((((((..(((((....)))))...)))))))...))))))..)))))...... ( -24.00, z-score = -0.85, R) >droSim1.chr4 608791 117 - 949497 ---AGUAUAUUUUGGGUGUUGCUUACAAUUCCAGUAUUAGGAUCAUUUCAUCGUCGUCUUCGAAAGCAUCGUGUAAGUUGAUAAGCUUCUGAUGGUGCAGUUGAUUCAUUAUAUCUAUUU ---.(((((..(((((.((((....))))))))).....((((((........(((....)))..(((((((..(((((....)))))...)))))))...))))))..)))))...... ( -23.60, z-score = -0.52, R) >droSec1.super_30 505737 117 - 664411 ---AGUAUAUUUUGGGUGUUGCUUACAAUUCCAGUAUUAGGAUCAUUUCAUCGUCGUCUUCGAAAGCAUCGUGUAAGUUGAUAAGCUUCUGAUGGUGCAGUUGAUUCAUUAUAUCUAUUU ---.(((((..(((((.((((....))))))))).....((((((........(((....)))..(((((((..(((((....)))))...)))))))...))))))..)))))...... ( -23.60, z-score = -0.52, R) >droYak2.chr4 853054 117 - 1374474 ---AGUAUAUUUUGGGUGUUACUUACAAUUCCAGUAUUAGGAUCAUUUCAUCGUCGUCUUCGAAAGCAUCGUGCAAGUUGAUAAGCUUCUGAUGGUGCAAUUGAUUCAUUAUAUCUAUUU ---.(((((..((((((((.....)))..))))).....((((((.(((((((((..........((.....))(((((....)))))..))))))).)).))))))..)))))...... ( -23.40, z-score = -0.69, R) >droEre2.scaffold_4512 813285 117 + 1286254 ---AGUAUAUUUUGGGCGUUGCUUACAAUUCCAGUAUUAGGAUCAUUUCAUCGUCGUCUUCGAAAGCAUCGUGCAAGUUGAUUAGCUUUGGAUGGUGUAGUUGAUUCAUUAUAUCUAUUU ---((.((((..((((((.((........(((.......)))......)).))))....((((..(((((((.(((((......)).))).)))))))..))))..))..)))))).... ( -23.34, z-score = -0.23, R) >droAna3.scaffold_13089 657183 106 - 978470 --------------UGAAUAACUUACAAUUCCAAUAUAAGAACCAUUUCGUCAUCAUCAUCAAAUGCGUCGUGCAAGUUAAUAAGCUUCUGAUGGUGUAGCUGAUUCAUAACAUCUAUUU --------------(((((....................(((....)))((...((((((((..(((.....)))((((....))))..))))))))..))..)))))............ ( -14.20, z-score = 0.51, R) >dp4.Unknown_group_8 17822 109 + 194412 -----------AUGGGGUUUGCUUACAAUUCUAGUAUUAGAACCAUUUCGUCGUCGUCUUCGAAGGCAUCGUGCAGAUUGAUAAGUUUUUGAUGGUGCAGCUGGUUCAUAAUAUCUAUUU -----------.(((((((.......)))))))(((((((((((((((((..(....)..)))))(((((((.((((..........)))))))))))...)))))).))))))...... ( -25.40, z-score = -0.91, R) >droPer1.super_42 592321 109 - 623919 -----------AUGGGGUUUGCUUACAAUUCUAGUAUUAGAACCAUUUCGUCGUCGUCUUCGAAGGCAUCGUGCAGAUUGAUAAGUUUUUGAUGGUGCAGCUGGUUCAUAAUAUCUAUUU -----------.(((((((.......)))))))(((((((((((((((((..(....)..)))))(((((((.((((..........)))))))))))...)))))).))))))...... ( -25.40, z-score = -0.91, R) >droWil1.scaffold_181130 1708081 117 - 16660200 --CGGUGUUUUUUCGGU-CUACUUACAAUUCCAGAAUAAGAACCAUUUCGUCAUCGUCUUCAAAUGCAUCGUGCAGAUUGAUUAGCUUCUGAUGGUGCAAUUGAUUCAUGAUAUCGAUCU --.(((.(((.(((((.-............)).))).))).)))...(((..(((((..((((.((((((((.((((..........)))))))))))).))))...)))))..)))... ( -29.82, z-score = -1.71, R) >droMoj3.scaffold_6498 593362 116 - 3408170 ----UAUGAAUUUUUCUUUUACUUACAAUUCAAGUAUAAGAACCAUUUCGUCAUCAUCUUCGAAAGCAUCGUGUAAGUUGAUCAAUUUUUGAUGAUGCAACUGGUUCAUAAUGUCAAUCU ----((((((...(((((.(((((.......))))).)))))((((((((..........)))))(((((((..(((((....)))))...)))))))...))))))))).......... ( -24.10, z-score = -2.07, R) >droVir3.scaffold_13052 1584201 117 - 2019633 ---GAUAAAUAUUAAGAUAUACUCACAAUUCUAGUAUGAGGACCAUUUCGUCAUCGUCUUCAAACGCAUCGUGUAGGUUUAUUAGUUUUUGGUGAUGUAACUGAUUCAUAAUGUCGAUCU ---(((..((((((.((((.((..(((......((.(((((((.((......)).))))))).))......)))..)).)))).....(..((......))..)....))))))..))). ( -22.80, z-score = -0.32, R) >droGri2.scaffold_14822 593084 110 - 1097346 ----------UUUCACUUGUACUCACAAUUCGAGAAUCAGGACCAUUUCGUCGUCGUCCUCGAAGGCGUCGUGCAGAUUGAUUAGUUUCUGGUGAUGCAGCUGAUUCAUGAUAUCGAUUU ----------..(((.(((((.((((........((((((...(((..((((.(((....))).))))..)))....))))))........))))))))).)))................ ( -24.99, z-score = 0.93, R) >anoGam1.chr2R 55570297 94 + 62725911 --------------------------AACUCGUAGAUCAGCACCAUCUCAUCCUCAUCCUCGAACGCGUCGUGCAGAUAGAUCAGCUUGCGGUGGUGCAGCUGGUUCAUGAUGUCGAUCU --------------------------.((((((..((((((((((((.((..((.(((.((..(((...)))...))..))).))..)).))))))...))))))..)))).))...... ( -25.30, z-score = 0.41, R) >apiMel3.Group2 14149948 120 + 14465785 AAUAUAUAUUAUACAACGACACUUACAACUCGAAGAUCAGAACCAUCUCGUCAUCGUCCUCGAACGCGUCGUGAAGAUUGAUCAAUUUCGGAUGAUGGAGUUGAUUCAUUAUGUCAAUUU .................((((.((((.((.((..(((.(((....))).))).(((....))).)).)).)))).(((.(((((((((((.....))))))))))).))).))))..... ( -24.80, z-score = 0.31, R) >triCas2.ChLG1X 1262621 94 - 8109244 --------------------------AACUCAUAGAUGAGGACCAUCUCGUCAUCAUCCUCAAAGGCAUCGUGCAAAUUGAUGAGUUUGGGGUGGUGGAGUUGGUUCAUGAUAUCGAUUU --------------------------........((((.((((((.(((..((((((((.((((..(((((.......)))))..))))))))))))))).))))))....))))..... ( -32.40, z-score = -2.57, R) >consensus _____U__AUUUUGGGUGUUACUUACAAUUCCAGUAUUAGGACCAUUUCGUCGUCGUCUUCGAAAGCAUCGUGCAAGUUGAUAAGCUUCUGAUGGUGCAGUUGAUUCAUUAUAUCUAUUU .......................................((((((.(((((((((..........((.....))(((((....)))))..))))))).)).))))))............. (-15.08 = -13.23 + -1.86)
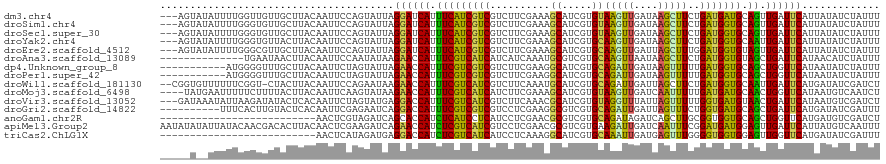

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:32 2011