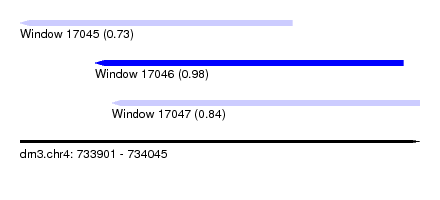
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 733,901 – 734,045 |
| Length | 144 |
| Max. P | 0.976180 |

| Location | 733,901 – 733,999 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Shannon entropy | 0.16216 |
| G+C content | 0.47271 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 733901 98 - 1351857 UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGUACGCCAAAGGAUACCUUU---------AGUAUGUGCAAUCAUCGAAAGUAUCACAAGCUUAGUCGCAUGUAAG ....(((....)))....((((((((((((((((((((.(((((...)))))---------.))..))))......(....).......))))).).)))))))).. ( -27.70, z-score = -1.78, R) >droEre2.scaffold_4512 755554 107 + 1286254 UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCACGCCAAAGGAAUCCUUUGCCAUGUGCACUAUGUGCACUCAUCGAAAGUAUCGCAAGCUUAGUCGCUUGUAAG (((((((((.....)..(((...((....))..))).))))))))....((((....((((((...))))))....))))......((((((......))))))... ( -34.50, z-score = -2.06, R) >droYak2.chr4 794802 98 - 1374474 UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCAAGCCAAAGGAAACCUUU---------ACUUUGUGCACUAAUCGAAAGUAUCGCAAGCAUAGUCGCAUGUAAG (((((((((.((.....)).)..((....))......)))))))).....((---------((..((((.((((..(....)...(....)..)))))))).)))). ( -26.70, z-score = -1.54, R) >droSec1.super_30 450284 98 - 664411 UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUU---------AGUAUGUGCACUCAUCGAAAGUAUCGCAAGCUUAGUCGCAUGUGAG (((((((((.....)..(((((.((....))))))).))))))))...((((---------.(.(((......)))).))))..(((((.((......)).))))). ( -32.40, z-score = -2.00, R) >droSim1.chr4 560585 98 - 949497 UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCGAAGGAUACCUUU---------AGUAUGUGCACUCAUCGAAAGUAUCGCAAGCUUAGUCGCAUGUAAG (((((((((.....)..(((((.((....))))))).)))))))).......---------..((((((((((...(....)...(....)...))).))))))).. ( -31.20, z-score = -1.74, R) >consensus UCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUU_________AGUAUGUGCACUCAUCGAAAGUAUCGCAAGCUUAGUCGCAUGUAAG (((((((((.....)..(((((.((....))))))).))))))))..................(((((((......(....)...(....).......))))))).. (-25.00 = -25.52 + 0.52)

| Location | 733,928 – 734,039 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Shannon entropy | 0.41390 |
| G+C content | 0.48921 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -24.54 |
| Energy contribution | -26.06 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 733928 111 - 1351857 AAUCCGGCACCAGUUCCCGCUUUCCUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGUACGCCAAAGGAUACCUUUAGUAUGUGCAAUCAUCGAA--------- ....(((...(((...........)))((((((((..((.(((((((((.....)..(((((.((....))))))).)))))))).)).........)))))))).)))..--------- ( -32.80, z-score = -1.29, R) >droEre2.scaffold_4512 755581 119 + 1286254 AAUCCGGAAACUGCGUCCGCUUU-CUGGAGAGCAUUCGCUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCACGCCAAAGGAAUCCUUUGCCAUGUGCACUAUGUGCACUCAUCGAA ..((((((((..((....)))))-)))))(.(((...(.((((((((((.....)..(((...((....))..))).))))))))).)...))))..((((((...))))))........ ( -40.70, z-score = -2.03, R) >droYak2.chr4 794829 111 - 1374474 AAUCCGGAAACUGAGCCCGCUUUACUGGAGAGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCAAGCCAAAGGAAACCUUUACUUUGUGCACUAAUCGAA--------- ..((.(....).))((.((((((.....)))))....((((((((((((.((.....)).)..((....))......))))))))))).........).))..........--------- ( -30.70, z-score = -0.90, R) >droSec1.super_30 450311 93 - 664411 ------------------AAUCCGGUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUUAGUAUGUGCACUCAUCGAA--------- ------------------....((((((..(((((..((.(((((((((.....)..(((((.((....))))))).)))))))).)).........))))).))))))..--------- ( -34.10, z-score = -3.03, R) >droSim1.chr4 560612 93 - 949497 ------------------AAUCCGGUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCGAAGGAUACCUUUAGUAUGUGCACUCAUCGAA--------- ------------------....((((((..(((((..((.(((((((((.....)..(((((.((....))))))).)))))))).)).........))))).))))))..--------- ( -33.80, z-score = -2.69, R) >consensus AAUCCGG_A_C_G___CCGCUUUGCUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUUAGUAUGUGCACUCAUCGAA_________ ...........................((((((((..((.(((((((((.....)..(((((.((....))))))).)))))))).)).........))))))))............... (-24.54 = -26.06 + 1.52)


| Location | 733,934 – 734,045 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.35080 |
| G+C content | 0.50838 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -24.77 |
| Energy contribution | -26.65 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 733934 111 - 1351857 UCGACCAAUCCGGCACCAGUUCCCGCUUUCCUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGUACGCCAAAGGAUACCUUUAGUAUGUGCAAUC--------- ..((.((((((((....(((....)))...))))))))...))((.(((((((((.....)..(((((.((....))))))).)))))))).)).................--------- ( -33.20, z-score = -1.45, R) >droEre2.scaffold_4512 755587 119 + 1286254 UGGACCAAUCCGGAAACUGCGUCCGCUUU-CUGGAGAGCAUUCGCUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCACGCCAAAGGAAUCCUUUGCCAUGUGCACUAUGUGCACUC ........((((((((..((....)))))-)))))(.(((...(.((((((((((.....)..(((...((....))..))).))))))))).)...))))..((((((...)))))).. ( -40.70, z-score = -1.77, R) >droYak2.chr4 794835 111 - 1374474 UGGACCAAUCCGGAAACUGAGCCCGCUUUACUGGAGAGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUUCAAGCCAAAGGAAACCUUUACUUUGUGCACUA--------- ((..((.....((((((.(((...(((((.....))))).)))))))))....))..))...(((((((((....))........(((((...))))).....))))))).--------- ( -32.60, z-score = -1.03, R) >droSec1.super_30 450317 93 - 664411 UGGACCAAUCCG------------------GUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUUAGUAUGUGCACUC--------- .(((....)))(------------------(((.(((((....((.(((((((((.....)..(((((.((....))))))).)))))))).)).....))).)).)))).--------- ( -32.10, z-score = -2.02, R) >droSim1.chr4 560618 93 - 949497 UGGACCAAUCCG------------------GUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCGAAGGAUACCUUUAGUAUGUGCACUC--------- .(((....)))(------------------(((.(((((....((.(((((((((.....)..(((((.((....))))))).)))))))).)).....))).)).)))).--------- ( -31.80, z-score = -1.78, R) >consensus UGGACCAAUCCGG_A_C_G___CCGCUUU_CUGGAUUGCAUUCGUUUCCUUUGGGGACAACAAGUGCAUGCGCAAGCUGCACGCCAAAGGAUACCUUUAGUAUGUGCACUC_________ ((.((((((((((.................)))))))).....((.(((((((((.....)..(((((.((....))))))).)))))))).)).........)).))............ (-24.77 = -26.65 + 1.88)

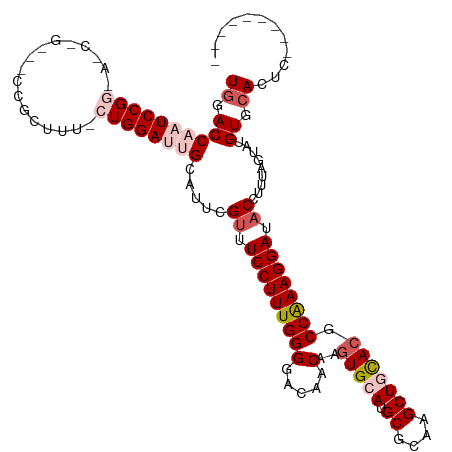
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:29 2011