
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 724,813 – 725,007 |
| Length | 194 |
| Max. P | 0.993718 |

| Location | 724,813 – 724,931 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.09 |
| Shannon entropy | 0.64112 |
| G+C content | 0.52440 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -16.38 |
| Energy contribution | -18.25 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 724813 118 + 1351857 CUGGGGGGGGGGGGGGUGUGGCACUGGGCGACUUCUCGAAAGGCGGUGAAAGCGGCAGGCGCUGCACCGUUUACACACAGUUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAG ((((.(((((..(..(((((((.....))...........((((((((..((((.....)))).)))))))).)))))..)..)))((((((.(.--.....).)))))).))...)))) ( -43.60, z-score = -0.81, R) >droAna3.scaffold_13099 2631239 94 - 3324263 -----------------------UCUAACAAAUAACCCUAA---GAAAAAAGUAAUAGGUAUUCCUAUCUAAAAAAAAACCUACAUACAUAAAUGUUGAUCGCAUGUGUUUAUAUUUUGG -----------------------..............((((---((........(((((....)))))................(((.(((.((((.....)))).))).))).)))))) ( -7.90, z-score = 0.54, R) >droEre2.scaffold_4512 746406 111 - 1286254 -------CUGGAAGAGUGUGAUGCCGGGCGCCUUUUCGGGAGGCGGUGAGAGCGGCAGCCGCUGCACCGUUUACAAGCAGCAUCUUACUGGAAGG--AAUAGCAUUCAGUUCCAGACCAA -------(((((((((((((((((...(((((((.....)))))((((..(((((...))))).))))........)).)))))...((....))--....))))))..))))))..... ( -42.30, z-score = -1.54, R) >droYak2.chr4 783105 111 + 1374474 -------AUGGAGGAGUGUGAGGCCGGGCGCCUUCUGGAGACGCGGUGAGCGCGGCAACCGCUGCACCGUUUAUAAACAGCAUCUCACGGGAAGG--AACAACAUUCAGUUCCAGAACAG -------.((((.((((((..(((.....)))(((((((((.((((((((((.(....))))).))))(((....))).)).)))).)))))...--....))))))...))))...... ( -41.50, z-score = -2.02, R) >droSec1.super_30 441762 111 + 664411 -------CUGGAGGAGUGUGGCACUGGGCGCCUUCUCGGAAGGCGGUAAAAGCGGUAGCUACUGCACCGUUUACACACAGCUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAG -------(((((((((((((........(((((((...)))))))((((..(((((.((....))))))))))).)))).))))..((((((.(.--.....).)))))))))))..... ( -44.40, z-score = -2.57, R) >droSim1.chr4 551742 111 + 949497 -------CUGGAGGAGUGUGGCACUGGGCGCCUUCUCGGAAGGCGGUGAAAGCGGUAGCUACUGCACCGUUUACACACAGCUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAG -------......(.((.(((.(((((((((((((...)))))))(((.(((((((.((....))))))))).))).((((.....)))).....--.......)))))).))).))).. ( -45.60, z-score = -2.55, R) >consensus _______CUGGAGGAGUGUGGCACCGGGCGCCUUCUCGGAAGGCGGUGAAAGCGGCAGCCACUGCACCGUUUACAAACAGCUUCUUACUGGAAGG__AACAACAUCCAGUUCCAGACCAG ..........((((((.(((........)))))))))...(((((((....(((((....))))))))))))..........(((.((((((.(........).))))))...))).... (-16.38 = -18.25 + 1.87)

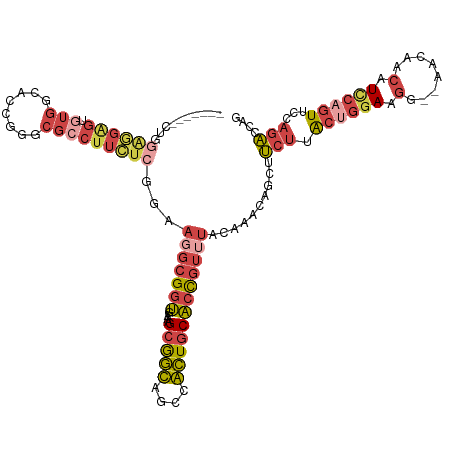

| Location | 724,853 – 724,967 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.62 |
| Shannon entropy | 0.59160 |
| G+C content | 0.46432 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 724853 114 + 1351857 AGGCGGUGAAAGCGGCAGGCGCUGCACCGUUUACACACAGUUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAGCACUUUCAUGU---CUGUUCAGGACCAAACCAUUCCAUU ((((((((..((((.....)))).))))))))(((.(((......((((((..((--(((........)))))...))))))......)))---.)))...(((.........)))... ( -35.50, z-score = -1.90, R) >droAna3.scaffold_13099 2631259 105 - 3324263 ------AAAAAGUAAUAGGUAUUCCUAUCUAAAAAAAAACCUACAUACAUAAAUGUUGAUCGCAUGUGUUUAUAUUUUGGCAUUGCUUUAC------UUC--CAUCUACCAUUCUCAAU ------...((((((((((....)))).............(.((((......)))).)...(((.(((((........)))))))).))))------)).--................. ( -11.90, z-score = -0.33, R) >droEre2.scaffold_4512 746439 117 - 1286254 AGGCGGUGAGAGCGGCAGCCGCUGCACCGUUUACAAGCAGCAUCUUACUGGAAGG--AAUAGCAUUCAGUUCCAGACCAAAUUUUCCAUGUAGACUGUUCAGAACGGAACCAUACUAGU ....(((....(((((....))))).((((((...(((((..(((.(((((((((--(((........)))))..........))))).))))))))))..)))))).)))........ ( -32.20, z-score = -1.02, R) >droYak2.chr4 783138 117 + 1374474 ACGCGGUGAGCGCGGCAACCGCUGCACCGUUUAUAAACAGCAUCUCACGGGAAGG--AACAACAUUCAGUUCCAGAACAGGAGUUCCUUGUAGACAGUUCAGGACGGAACCAUACCACU ..((((((((((.(....))))).))))))............(((.(((((..((--(((........))))).((((....))))))))))))..((((......))))......... ( -34.50, z-score = -1.84, R) >droSec1.super_30 441795 117 + 664411 AGGCGGUAAAAGCGGUAGCUACUGCACCGUUUACACACAGCUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAGCAGUUUCAUGUAGACAGUUCAGGACGAAACCAUUCCAGU .((.(((...(((....))).(((.((.(((((((...((((....(((((..((--(((........)))))...)))))))))...))))))).)).)))......)))...))... ( -33.90, z-score = -2.08, R) >droSim1.chr4 551775 117 + 949497 AGGCGGUGAAAGCGGUAGCUACUGCACCGUUUACACACAGCUUCUUGCUGGAAGG--AACAACAUCCAGUUCCAGACCAGCAGUUUCAUGUAGACAGUUCAGGACGAAACCAUUCCAGU ((((.(((.(((((((.((....))))))))).)))...))))..((((((..((--(((........)))))...))))))(((((.((....))(((...))))))))......... ( -37.30, z-score = -2.57, R) >consensus AGGCGGUGAAAGCGGCAGCCACUGCACCGUUUACAAACAGCUUCUUACUGGAAGG__AACAACAUCCAGUUCCAGACCAGCAGUUCCAUGUAGACAGUUCAGGACGAAACCAUUCCAGU (((((((....(((((....))))))))))))..........(((.((((((.(........).))))))...)))........................................... (-12.65 = -13.43 + 0.78)

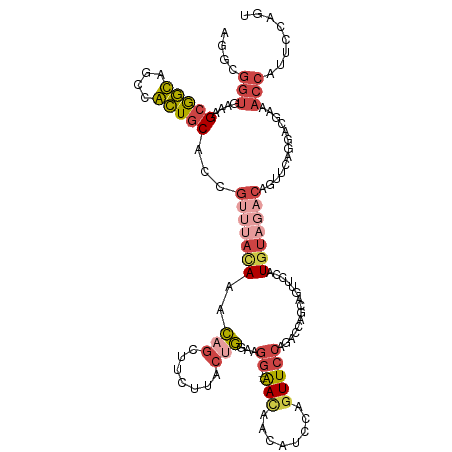

| Location | 724,893 – 725,007 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.29308 |
| G+C content | 0.47130 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -27.18 |
| Energy contribution | -32.06 |
| Covariance contribution | 4.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 724893 114 - 1351857 AAUAAAGGAAGAACCAACUCUAGACGGACCAGCAGCACAUAAUGGAAUGGUUUGGUCCUGAACAG---ACAUGAAAGUGCUGGUCUGGAACUGGAUGUUGUUCCUUCCAGCAAGAAA ......(((((...(((((((((.((((((((((...(((......)))(((((........)))---)).......))))))))))...))))).))))...)))))......... ( -39.90, z-score = -3.52, R) >droEre2.scaffold_4512 746479 117 + 1286254 AAUAAAGGACGGCCCAACACCAGCCGCACCAGCAGCACAUACUAGUAUGGUUCCGUUCUGAACAGUCUACAUGGAAAAUUUGGUCUGGAACUGAAUGCUAUUCCUUCCAGUAAGAUG ......(((.((..........((.((....)).))......(((((((((((((..(..((...(((....)))...))..)..)))))))..))))))..)).)))......... ( -27.30, z-score = -0.24, R) >droYak2.chr4 783178 112 - 1374474 AAUAAAGG-----CCAAAUGCAAACGCUCCAGCAGCACAUAGUGGUAUGGUUCCGUCCUGAACUGUCUACAAGGAACUCCUGUUCUGGAACUGAAUGUUGUUCCUUCCCGUGAGAUG ....((((-----.(((..((....))(((((.(((.....((((.(((((((......))))))))))).(((....)))))))))))........)))..))))..(....)... ( -25.80, z-score = 0.01, R) >droSec1.super_30 441835 117 - 664411 AAUAAAGGAAGGACCGACUCCAGACGGACCAGCAGCACAUACUGGAAUGGUUUCGUCCUGAACUGUCUACAUGAAACUGCUGGUCUGGAACUGGAUGUUGUUCCUUCCAGCAAGAAG ......(((((((.(((((((((.(((((((((((..(((..((((.((((((......)))))))))).)))...)))))))))))...))))).)))).)))))))......... ( -51.00, z-score = -5.81, R) >droSim1.chr4 551815 117 - 949497 AAUAAAGGAAGGACCGACUCCAGACGGACCAGCAGCACAUACUGGAAUGGUUUCGUCCUGAACUGUCUACAUGAAACUGCUGGUCUGGAACUGGAUGUUGUUCCUUCCAGCAAGAAG ......(((((((.(((((((((.(((((((((((..(((..((((.((((((......)))))))))).)))...)))))))))))...))))).)))).)))))))......... ( -51.00, z-score = -5.81, R) >consensus AAUAAAGGAAGGACCAACUCCAGACGGACCAGCAGCACAUACUGGAAUGGUUUCGUCCUGAACUGUCUACAUGAAACUGCUGGUCUGGAACUGGAUGUUGUUCCUUCCAGCAAGAAG ......(((((((.(((((((((.(((((((((((.......(((.(((((((......)))))))))).......)))))))))))...))))).)))).)))))))......... (-27.18 = -32.06 + 4.88)


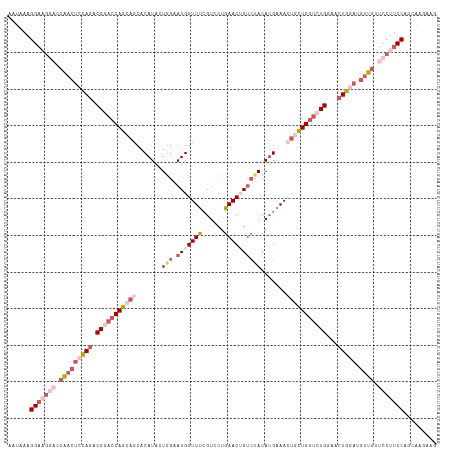
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:27 2011