
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 723,686 – 723,785 |
| Length | 99 |
| Max. P | 0.790297 |

| Location | 723,686 – 723,785 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.57178 |
| G+C content | 0.44268 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -12.85 |
| Energy contribution | -10.28 |
| Covariance contribution | -2.58 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 723686 99 - 1351857 ------GCCUAAAGGUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUGAAAGUAUGUCC---CCCGCAGGAAGCUG-AAUCAGCCCUGCUAUCAUCAUA ------(((....)))((.(((.....((((((((((...(((((...)))))..)))))))))).)))---.))(((((..((((-...))))))))).......... ( -28.80, z-score = -1.57, R) >droSim1.chr4 550559 99 - 949497 ------GCCUGAAGGUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUGAAAGUAUGUCC---CCCGCAGGAAGCUG-AAUCAGCCCUGCAAUCAUCAUA ------...(((.(((((.(((.....((((((((((...(((((...)))))..)))))))))).)))---.))(((((..((((-...))))))))).))).))).. ( -29.60, z-score = -1.65, R) >droSec1.super_30 440607 99 - 664411 ------GCCUGAAGGUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUGAAAGUAUGUCC---CCCGCAGGAAGCUG-AAUCAGCCUUGCAAUCAUCAUA ------...(((.(((((.(((.....((((((((((...(((((...)))))..)))))))))).)))---.))(((((..((((-...))))))))).))).))).. ( -26.90, z-score = -0.89, R) >droYak2.chr4 781534 99 - 1374474 ------GCCUGGAGAUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUAAAAGCUUGUCC---CCCGCAGGAAGCUG-AAUCUGUCUUGCAAUCAUCAUA ------.......(((((.((..((((..((((((((...(((((...)))))..))))))))))))..---)).((((((((...-...)).))))))..)))))... ( -26.30, z-score = -0.77, R) >droEre2.scaffold_4512 744962 99 + 1286254 ------GCCUGAAGGUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUGAAAGCUAGUUC---CCCGCAGGAAGCUG-AAUCAGCCUUGCAAUCAUCAUA ------...(((.(((((.((((......((((((((...(((((...)))))..))))))))...)))---)))(((((..((((-...))))))))).))).))).. ( -29.70, z-score = -1.66, R) >droAna3.scaffold_13099 2627201 104 + 3324263 -GUAUUAUACGUAUCUUCACGA---AAAUGUUUAUGCCACAUGGCAGCACAUUAAAGAAAAUCAAAUUCCUUUUCGAAGGAAGUGG-ACGCCGCUUUGCAAUCAUCACA -.........((..((((....---.(((((...((((....))))..)))))...(((((.........)))))))))(((((((-...))))))))).......... ( -21.00, z-score = -1.10, R) >dp4.Unknown_group_10 25290 107 - 72859 UGUACGGUGCGGGGGUGGAAGUUGCGAAAUUUCUUAUUGUAUUGUAUGGGAGCGACUGAAAGUAAAUCC--AUCGGCAGGAAGUAGAAACCAGUCCUGCAAUCUGCACA ......(((((((((((((((((((......((((((........)))))))))))).........)))--))).((((((.((....))...))))))..))))))). ( -34.60, z-score = -2.32, R) >droPer1.super_42 528224 107 - 623919 UGUACGGUGCGGGGGUGGAAGUUGCGAAAUUUCUUAUUGUAUUGUAUGGGAGCGACUGAAAGUAAAUCC--AUCGGCAGGAAGUAGAAACCAGUCCUGCAAUCUGCACA ......(((((((((((((((((((......((((((........)))))))))))).........)))--))).((((((.((....))...))))))..))))))). ( -34.60, z-score = -2.32, R) >consensus ______GCCUGAAGGUGGAGGAAACAAAUGUUUUUAUUAGGCUACAUGGUAGCACAUGAAAGUAAGUCC___CCCGCAGGAAGCUG_AAUCAGCCCUGCAAUCAUCAUA ......((((((....((((.(......).))))..)))))).................................(((((..((((....))))))))).......... (-12.85 = -10.28 + -2.58)
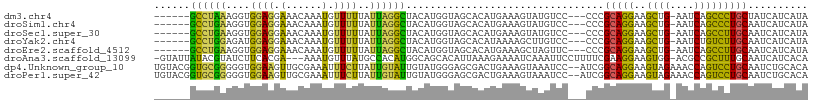
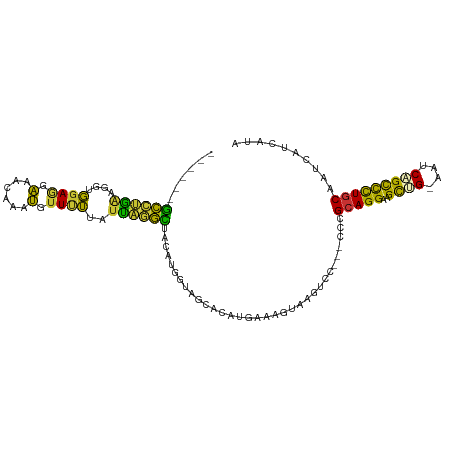
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:24 2011