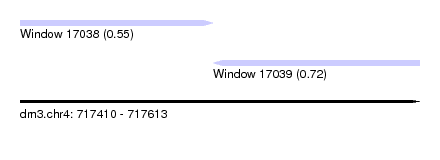
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 717,410 – 717,613 |
| Length | 203 |
| Max. P | 0.718878 |

| Location | 717,410 – 717,508 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 72.38 |
| Shannon entropy | 0.44665 |
| G+C content | 0.35888 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -11.26 |
| Energy contribution | -12.32 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 717410 98 + 1351857 UAAUAUGGUUGUGUAAGAAGGCAAUCUGGGAACUCCAUAAAAAGGGUAGGGACAAUUACCGCGAUGUCUAGUGUUUUCCAGAUUAGCUGUCGAUAUAU ..........(((((.((.(((((((((((((((((........)).))(((((..........))))).....)))))))))).))).))..))))) ( -23.50, z-score = -0.95, R) >droYak2.chr4 770351 83 + 1374474 -------------UAAAAUGCU--UUUGGAAACUCCAUAAUUAGGUCGCUGACUAUUCACGCGAUACGUAGUGUUUUCCUUAUUAGUCAUCGAUAUAU -------------.........--..(((.....))).......((((.((((((...((((........)))).........)))))).)))).... ( -14.30, z-score = -0.53, R) >droSec1.super_30 433989 93 + 664411 UAAAAAAGUUUUGUAAGAAGGCAAUCUGGGAACUCCAUAAAUAGGGGAGGGAUAAUUACCACGAUAUUU-----UUUCCAGAUAAGCUGUCGAUAUAU ...........((((.((.(((.(((((((((((((.......)))).((........)).........-----)))))))))..))).))..)))). ( -21.60, z-score = -1.94, R) >droSim1.chr4 544062 93 + 949497 UAAAAAAGUUUUGUAAGAAGGCAAUCUGGGAACUCCAUAAAUAGGGGAGGGAUAAUUACCACGAUAUUU-----UUUCCAGAUAAGCUGUCGAUAUAU ...........((((.((.(((.(((((((((((((.......)))).((........)).........-----)))))))))..))).))..)))). ( -21.60, z-score = -1.94, R) >consensus UAAAAAAGUUUUGUAAGAAGGCAAUCUGGGAACUCCAUAAAUAGGGGAGGGACAAUUACCACGAUAUUU_____UUUCCAGAUAAGCUGUCGAUAUAU ................((.(((..((((((((((((.......)))).((........))..............))))))))...))).))....... (-11.26 = -12.32 + 1.06)

| Location | 717,508 – 717,613 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.50898 |
| G+C content | 0.36587 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -11.54 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 717508 105 - 1351857 -AUAAAAAUAAGUUAGUACAGUGUGAACACCUAUCGUUUUCUUCCA------UAUCGAUAGGAAACUCAUAAAUGUAUGUGUGGAAAGGCUUAAACAUCUGUACGGGCGUCG -..........((..(((((((((((...(((((((..........------...)))))))....))))).((((..((.(....).))....))))))))))..)).... ( -24.22, z-score = -1.70, R) >droEre2.scaffold_4512 738381 111 + 1286254 -AACACAAUAUGUUAGUCCAGUGUGAACAUCUAUUCUGCGUUUUCUUUUCCUUAUCGAUAGGAAACUCACGUAUUUAUGUGUGAAAAGGCUUUAACAUCCGUAUUAACGUCG -........(((((((((..(((((((......))).((.(((((.((((((.......))))))..(((((....))))).))))).))....))))..).)))))))).. ( -21.30, z-score = -1.05, R) >droYak2.chr4 770434 109 - 1374474 -AAAACAAUAAGUUAGAUCAGUGUGAACACCUUUUCUGCGGUUUGUUUUA--UAUCGAUAAUAAAGUCACGAAUGUAUGUGUGAAAAGGCUUUAACAUCAGUAUUAACGUCG -(((((((...((.(((...(((....)))....)))))...)))))))(--(((.(((..((((((((((........)))).....))))))..))).))))........ ( -17.60, z-score = 0.52, R) >droSec1.super_30 434082 85 - 664411 --CUAAAAAAAGUUUGUACAGUGUGAAAACCU-------------------------AUAGGAAACUGAUAAAUGCAUGUGUGGAAAGGCUUUAACAUUCGUACGGACGUCG --.........(((((((((((((.(((.(((-------------------------.(((....)))......((....))....))).))).))))).)))))))).... ( -20.50, z-score = -2.41, R) >droSim1.chr4 544155 87 - 949497 CUAAAAAAAAAGUUUGUACAGUGUGAACACCU-------------------------AUAGGAAACUGAUAAAUGCAUGUGUGGAAAGGCUUUAACAUCCGUACGGACGUCG ...........((((((((.(((....)))..-------------------------.(((....)))......(.((((..((.....))...)))).))))))))).... ( -18.50, z-score = -1.23, R) >consensus _AAAAAAAUAAGUUAGUACAGUGUGAACACCU_U__U____UU_________UAUCGAUAGGAAACUCAUAAAUGUAUGUGUGGAAAGGCUUUAACAUCCGUACGGACGUCG ...........((((((((.((((.....(((............................(....)(((((........)))))..))).....))))..)))))))).... (-11.54 = -11.10 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:23 2011